Cannabinoid Receptor Type-2 in B Cells Is Associated with Tumor Immunity in Melanoma
Abstract
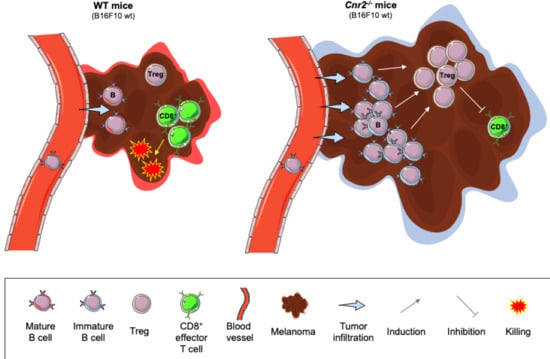
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Culture
2.2. Mice, Tumor Inoculation and In Vivo Studies
2.3. Flow Cytometric Analyses and Cell Sorting
2.4. Fluorescence In Situ Hybridization (FISH) Using RNAscope
2.5. Single-Cell RNA Sequencing
2.6. TCGA Data Collection and Analysis
2.7. Analysis of Gene Expression Omnibus (GEO) Dataset
2.8. Statistical Analysis
3. Results
3.1. B Cell-Associated CB2Rs Play a Role in Human Melanoma Progression
3.2. Cnr2 Deficiency Leads to Enhanced Melanoma Growth and B Cell Infiltration in Murine Melanoma
3.3. Tumor-Infiltrating B Cells of Cnr2-Defficient Mice Display Signs of Impaired Differentiation and Catabolism
3.4. Cnr2-Deficient B Cells Drive the Induction of Treg
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muir, R. Melanoma. N. Engl. J. Med. 2006, 355, 51–65. [Google Scholar]
- Rebecca, V.W.; Somasundaram, R.; Herlyn, M. Pre-clinical modeling of cutaneous melanoma. Nat. Commun. 2020, 11, 2858. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glazer, A.M.; Winkelmann, R.R.; Farberg, A.S.; Rigel, D.S. Analysis of Trends in US Melanoma Incidence and Mortality. JAMA Dermatol. 2017, 153, 225–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadozai, H.; Gruber, T.; Hunger, R.E.; Schenk, M. Recent Successes and Future Directions in Immunotherapy of Cutaneous Melanoma. Front. Immunol. 2017, 8, 1617. [Google Scholar] [CrossRef]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef] [Green Version]
- Moreno, E.; Cavic, M.; Krivokuca, A.; Casadó, V.; Canela, E. The Endocannabinoid System as a Target in Cancer Diseases: Are We There Yet? Front. Pharmacol. 2019, 10, 339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladin, D.A.; Soliman, E.; Griffin, L.; Van Dross-Anderson, R. Preclinical and Clinical Assessment of Cannabinoids as Anti-Cancer Agents. Front. Pharmacol. 2016, 7, 361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velasco, G.; Sánchez, C.; Guzmán, M. Towards the use of cannabinoids as antitumour agents. Nat. Rev. Cancer 2012, 12, 436–444. [Google Scholar] [CrossRef]
- Sugiura, T.; Kondo, S.; Sukagawa, A.; Nakane, S.; Shinoda, A.; Itoh, K.; Yamashita, A.; Waku, K. 2-arachidonoylglycerol: A possible endogenous cannabinoid receptor ligand in brain. Biochem. Biophys. Res. Commun. 1995, 215, 89–97. [Google Scholar] [CrossRef]
- Devane, W.A.; Hanus, L.; Breuer, A.; Pertwee, R.G.; Stevenson, L.A.; Griffin, G.; Gibson, D.; Mandelbaum, A.; Etinger, A.; Mechoulam, R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 1992, 258, 1946–1949. [Google Scholar] [CrossRef]
- Mechoulam, R.; Ben-Shabat, S.; Hanus, L.; Ligumsky, M.; Kaminski, N.E.; Schatz, A.R.; Gopher, A.; Almog, S.; Martin, B.R.; Compton, D.R.; et al. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem. Pharmacol. 1995, 50, 83–90. [Google Scholar] [CrossRef]
- Matsuda, L.A.; Lolait, S.J.; Brownstein, M.J.; Young, A.C.; Bonner, T.I. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature 1990, 346, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.; Thomas, K.L.; Abu-Shaar, M. Molecular characterization of a peripheral receptor for cannabinoids. Nature 1993, 365, 61–65. [Google Scholar] [CrossRef]
- Galiegue, S.; Mary, S.; Marchand, J.; Dussossoy, D.; Carriere, D.; Carayon, P.; Bouaboula, M.; Shire, D.; Fur, G.; Casellas, P. Expression of Central and Peripheral Cannabinoid Receptors in Human Immune Tissues and Leukocyte Subpopulations. J. Biol. Inorg. Chem. 1995, 232, 54–61. [Google Scholar] [CrossRef]
- Ziring, D.; Wei, B.; Velazquez, P.; Schrage, M.; Buckley, N.E.; Braun, J. Formation of B and T cell subsets require the cannabinoid receptor CB2. Immunogenetics 2006, 58, 714–725. [Google Scholar] [CrossRef]
- Pereira, J.P.; An, J.; Xu, Y.; Huang, Y.; Cyster, J.G. Cannabinoid receptor 2 mediates the retention of immature B cells in bone marrow sinusoids. Nat. Immunol. 2009, 10, 403–411. [Google Scholar] [CrossRef]
- Kienzl, M.; Kargl, J.; Schicho, R. The Immune Endocannabinoid System of the Tumor Microenvironment. Int. J. Mol. Sci. 2020, 21, 8929. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, R.; Zhang, G.; Fukunaga-Kalabis, M.; Perego, M.; Krepler, C.; Xu, X.; Wagner, C.; Hristova, D.; Zhang, J.; Tian, T.; et al. Tumor-associated B-cells induce tumor heterogeneity and therapy resistance. Nat. Commun. 2017, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erdag, G.; Schaefer, J.T.; Smolkin, M.E.; Deacon, D.H.; Shea, S.M.; Dengel, L.T.; Patterson, J.W.; Slingluff, C.L. Immunotype and Immunohistologic Characteristics of Tumor-Infiltrating Immune Cells Are Associated with Clinical Outcome in Metastatic Melanoma. Cancer Res. 2012, 72, 1070–1080. [Google Scholar] [CrossRef] [Green Version]
- Yuen, G.J.; Demissie, E.; Pillai, S. B Lymphocytes and Cancer: A Love–Hate Relationship. Trends Cancer 2016, 2, 747–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakaguchi, S. Regulatory T cells: Key controllers of immunologic self-tolerance. Cell 2000, 101, 455–458. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Sakaguchi, S. Targeting Treg cells in cancer immunotherapy. Eur. J. Immunol. 2019, 49, 1140–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joosten, M. Leukemic predisposition of pSca-1/Cb2 transgenic mice. Exp. Hematol. 2002, 30, 142–149. [Google Scholar] [CrossRef]
- Zheng, D.; Bode, A.M.; Zhao, Q.; Cho, Y.-Y.; Zhu, F.; Ma, W.-Y.; Dong, Z. The Cannabinoid Receptors Are Required for Ultraviolet-Induced Inflammation and Skin Cancer Development. Cancer Res. 2008, 68, 3992–3998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Martínez, E.; Martín-Ruiz, A.; Martín, P.; Calvo, V.; Provencio, M.; García, J.M. CB2 cannabinoid receptor activation promotes colon cancer progression via AKT/GSK3β signaling pathway. Oncotarget 2016, 7, 68781–68791. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Ma, H.; Bo, Y.; Shao, M. The oncogenic role of CB2 in the progression of non-small-cell lung cancer. Biomed. Pharmacother. 2019, 117, 109080. [Google Scholar] [CrossRef]
- Carmi, Y.; Spitzer, M.H.; Linde, I.L.; Burt, B.M.; Prestwood, T.R.; Perlman, N.; Davidson, M.G.; Kenkel, J.A.; Segal, E.; Pusapati, G.V.; et al. Allogeneic IgG combined with dendritic cell stimuli induce antitumour T-cell immunity. Nature 2015, 521, 99–104. [Google Scholar] [CrossRef]
- DeMaria, O.; De Gassart, A.; Coso, S.; Gestermann, N.; Di Domizio, J.; Flatz, L.; Gaide, O.; Michielin, O.; Hwu, P.; Petrova, T.V.; et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc. Natl. Acad. Sci. USA 2015, 112, 15408–15413. [Google Scholar] [CrossRef] [Green Version]
- Gruber, T.; Kremenovic, M.; Sadozai, H.; Rombini, N.; Baeriswyl, L.; Maibach, F.; Modlin, R.L.; Gilliet, M.; Von Werdt, D.; Hunger, R.E.; et al. IL-32γ potentiates tumor immunity in melanoma. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Qu, H.; Wang, S.; Wei, J.; Zhang, L.; Ma, R.; Lu, J.; Zhu, J.; Zhong, W.-D.; Jia, Z. GDCRNATools: An R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics 2018, 34, 2515–2517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Hastie, T. Regularized paths for generalized linear models via coordinate descent (Technical Report). J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [Green Version]
- Soethoudt, M.; Grether, U.; Fingerle, J.; Grim, T.W.; Fezza, F.; De Petrocellis, L.; Ullmer, C.; Rothenhäusler, B.; Perret, C.; Van Gils, N.; et al. Cannabinoid CB2 receptor ligand profiling reveals biased signalling and off-target activity. Nat. Commun. 2017, 8, 13958. [Google Scholar] [CrossRef]
- Casanova, M.L.; Blázquez, C.; Martínez-Palacio, J.; Villanueva, C.; Fernández-Aceñero, M.J.; Huffman, J.W.; Jorcano, J.L.; Guzmán, M. Inhibition of skin tumor growth and angiogenesis in vivo by activation of cannabinoid receptors. J. Clin. Investig. 2003, 111, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Javid, F.A.; Phillips, R.M.; Afshinjavid, S.; Verde, R.; Ligresti, A. Cannabinoid pharmacology in cancer research: A new hope for cancer patients? Eur. J. Pharmacol. 2016, 775, 1–14. [Google Scholar] [CrossRef]
- Belardelli, F.; Ferrantini, M.; Proietti, E.; Kirkwood, J.M. Interferon-alpha in tumor immunity and immunotherapy. Cytokine Growth Factor Rev. 2002, 13, 119–134. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Sarvaria, A.; Madrigal, J.A.; Saudemont, A. B cell regulation in cancer and anti-tumor immunity. Cell. Mol. Immunol. 2017, 14, 662–674. [Google Scholar] [CrossRef] [Green Version]
- Blázquez, C.; Carracedo, A.; Barrado, L.; Real, P.J.; Fernández-Luna, J.L.; Velasco, G.; Malumbres, M.; Guzmán, M. Cannabinoid receptors as novel targets for the treatment of melanoma. FASEB J. 2006, 20, E2199–E2208. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Yang, J.; Zhao, H.; Fang, X.; Li, H. Cannabinoid receptor 2 is upregulated in melanoma. J. Cancer Res. Ther. 2012, 8, 549–554. [Google Scholar] [CrossRef]
- Basu, S.; Ray, A.; Dittel, B.N. Cannabinoid Receptor 2 Is Critical for the Homing and Retention of Marginal Zone B Lineage Cells and for Efficient T-Independent Immune Responses. J. Immunol. 2011, 187, 5720–5732. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gruber, T.; Robatel, S.; Kremenovic, M.; Bäriswyl, L.; Gertsch, J.; Schenk, M. Cannabinoid Receptor Type-2 in B Cells Is Associated with Tumor Immunity in Melanoma. Cancers 2021, 13, 1934. https://doi.org/10.3390/cancers13081934
Gruber T, Robatel S, Kremenovic M, Bäriswyl L, Gertsch J, Schenk M. Cannabinoid Receptor Type-2 in B Cells Is Associated with Tumor Immunity in Melanoma. Cancers. 2021; 13(8):1934. https://doi.org/10.3390/cancers13081934
Chicago/Turabian StyleGruber, Thomas, Steve Robatel, Mirela Kremenovic, Lukas Bäriswyl, Jürg Gertsch, and Mirjam Schenk. 2021. "Cannabinoid Receptor Type-2 in B Cells Is Associated with Tumor Immunity in Melanoma" Cancers 13, no. 8: 1934. https://doi.org/10.3390/cancers13081934