Current Status and Perspectives of Patient-Derived Models for Ewing’s Sarcoma
Abstract
:Simple Summary
Abstract
1. Introduction
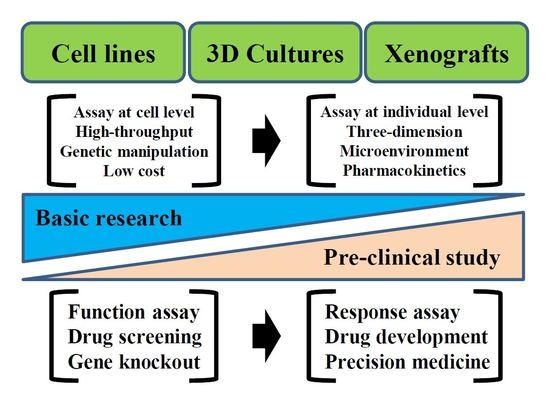
Ewing’s Sarcoma and Patient-Derived Cancer Models
2. Materials and Methods
3. Results: Technical Variation of Patient-Derived Cancer Models Used for Research on Ewing’s Sarcoma
3.1. Cell Lines
3.2. Organoids
3.3. PDXs
4. Conclusions
Future Perspectives of Patient-Derived Ewing’s Sarcoma Models
Funding
Conflicts of Interest
References
- Grier, H.E. The Ewing Family of Tumors. Pediatr. Clin. N. Am. 1997, 44, 991–1004. [Google Scholar] [CrossRef]
- Rodriguez-Galindo, C.; Liu, T.; Krasin, M.J.; Wu, J.; Billups, C.A.; Daw, N.C.; Spunt, S.L.; Rao, B.N.; Santana, V.M.; Navid, F. Analysis of prognostic factors in Ewing sarcoma family of tumors. Cancer 2007, 110, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Randall, R.L.; Lessnick, S.L.; Jones, K.B.; Gouw, L.G.; Cummings, J.E.; Albright, L.C.; Schiffman, J.D. Is There a Predisposition Gene for Ewing’s Sarcoma? J. Oncol. 2010, 2010, 397632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balamuth, N.J.; Womer, R.B. Ewing’s sarcoma. Lancet Oncol. 2010, 11, 184–192. [Google Scholar] [CrossRef]
- Delattre, O.; Zucman-Rossi, J.; Plougastel-Douglas, B.; Desmaze, C.; Melot, T.; Peter, M.; Kovar, H.; Joubert, I.; De Jong, P.; Rouleau, G.; et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992, 359, 162–165. [Google Scholar] [CrossRef]
- Dubois, S.G.; Grier, H.E.; Lessnick, S.L. Ewing’s Sarcoma. In Oncology of Infancy and Childhood, 1st ed.; Saunders: Philadelphia, PA, USA, 2009; pp. 829–869. [Google Scholar]
- Üren, A.; Toretsky, J.A. Ewing’s sarcoma oncoprotein EWS–FLI1: The perfect target without a therapeutic agent. Futur. Oncol. 2005, 1, 521–528. [Google Scholar] [CrossRef]
- Staege, M.S.; Hutter, C.; Neumann, I.; Foja, S.; Hattenhorst, U.E.; Hansen, G.; Afar, D.; Burdach, S.E.G. DNA Microarrays Reveal Relationship of Ewing Family Tumors to Both Endothelial and Fetal Neural Crest-Derived Cells and Define Novel Targets. Cancer Res. 2004, 64, 8213–8221. [Google Scholar] [CrossRef] [Green Version]
- Sheffield, N.C.; Pierron, G.; Klughammer, J.; Datlinger, P.; Schönegger, A.; Schuster, M.; Hadler, J.; Surdez, D.; Guillemot, D.; Lapouble, E.; et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 2017, 23, 386–395. [Google Scholar] [CrossRef]
- Brohl, A.S.; Solomon, D.A.; Chang, W.; Wang, J.; Song, Y.; Sindiri, S.; Patidar, R.; Hurd, L.; Chen, L.; Shern, J.F.; et al. The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent STAG2 Mutation. PLoS Genet. 2014, 10, e1004475. [Google Scholar] [CrossRef] [Green Version]
- Crompton, B.D.; Stewart, C.; Taylor-Weiner, A.; Alexe, G.; Kurek, K.C.; Calicchio, M.L.; Kiezun, A.; Carter, S.L.; Shukla, S.A.; Mehta, S.S.; et al. The Genomic Landscape of Pediatric Ewing Sarcoma. Cancer Discov. 2014, 4, 1326–1341. [Google Scholar] [CrossRef] [Green Version]
- Tirode, F.; Surdez, D.; Ma, X.; Parker, M.D.; Le Deley, M.-C.; Bahrami, A.; Zhang, Z.; Lapouble, E.; Grossetête-Lalami, S.; Rusch, M.; et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014, 4, 1342–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damron, T.; Ward, W.G.; Stewart, A. Osteosarcoma, Chondrosarcoma, and Ewing Sarcoma. Clin. Orthop. Relat. Res. 2007, 459, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Esiashvili, N.; Goodman, M.; Marcus, R.B. Changes in Incidence and Survival of Ewing Sarcoma Patients Over the Past 3 Decades. J. Pediatr. Hematol. 2008, 30, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Cotterill, S.; Ahrens, S.; Paulussen, M.; Jürgens, H.; Voûte, P.; Gadner, H.; Craft, A. Prognostic Factors in Ewing’s Tumor of Bone: Analysis of 975 Patients From the European Intergroup Cooperative Ewing’s Sarcoma Study Group. J. Clin. Oncol. 2000, 18, 3108–3114. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, S.; Hoffmann, C.; Jabar, S.; Braun-Munzinger, G.; Paulussen, M.; Dunst, J.; Rübe, C.; Winkelmann, W.; Heinecke, A.; Göbel, U.; et al. Evaluation of prognostic factors in a tumor volume-adapted treatment strategy for localized Ewing sarcoma of bone: The CESS 86 experience. Med Pediatr. Oncol. 1999, 32, 186–195. [Google Scholar] [CrossRef]
- Gaspar, N.; Hawkins, D.S.; Dirksen, U.; Lewis, I.J.; Ferrari, S.; Le Deley, M.-C.; Kovar, H.; Grimer, R.J.; Whelan, J.; Claude, L.; et al. Ewing Sarcoma: Current Management and Future Approaches Through Collaboration. J. Clin. Oncol. 2015, 33, 3036–3046. [Google Scholar] [CrossRef]
- Werier, J.; Yao, X.; Caudrelier, J.-M.; Di Primio, G.; Ghert, M.; Gupta, A.A.; Kandel, R.; Verma, S. A systematic review of optimal treatment strategies for localized Ewing’s sarcoma of bone after neo-adjuvant chemotherapy. Surg. Oncol. 2016, 25, 16–23. [Google Scholar] [CrossRef]
- Olmos, D.; Postel-Vinay, S.; Molife, L.R.; Okuno, S.H.; Schuetze, S.M.; Paccagnella, M.L.; Batzel, G.N.; Yin, D.; Pritchard-Jones, K.; Judson, I.; et al. Safety, pharmacokinetics, and preliminary activity of the anti-IGF-1R antibody figitumumab (CP-751,871) in patients with sarcoma and Ewing’s sarcoma: A phase 1 expansion cohort study. Lancet Oncol. 2010, 11, 129–135. [Google Scholar] [CrossRef] [Green Version]
- Manara, M.C.; Landuzzi, L.; Nanni, P.; Nicoletti, G.; Zambelli, D.; Lollini, P.L.; Nanni, C.; Hofmann, F.; Garcia-Echeverria, C.; Picci, P.; et al. Preclinical In vivo Study of New Insulin-Like Growth Factor-I Receptor-Specific Inhibitor in Ewing’s Sarcoma. Clin. Cancer Res. 2007, 13, 1322–1330. [Google Scholar] [CrossRef] [Green Version]
- Benini, S.; Manara, M.C.; Baldini, N.; Cerisano, V.; Serra, M.; Mercuri, M.; Lollini, P.L.; Nanni, P.; Picci, P.; Scotlandi, K. Inhibition of insulin-like growth factor I receptor increases the antitumor activity of doxorubicin and vincristine against Ewing’s sarcoma cells. Clin. Cancer Res. 2001, 7, 1790–1797. [Google Scholar]
- Mancarella, C.; Scotlandi, K. 40 YEARS OF IGF1: IGF system in sarcomas: A crucial pathway with many unknowns to exploit for therapy. J. Mol. Endocrinol. 2018, 61, T45–T60. [Google Scholar] [CrossRef] [PubMed]
- AACR Project GENIE Consortium. The AACR Project GENIE Consortium AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corsello, S.M.; Bittker, J.A.; Liu, Z.; Gould, J.; McCarren, P.; Hirschman, J.E.; Johnston, S.E.; Vrcic, A.; Wong, B.; Khan, M.; et al. The Drug Repurposing Hub: A next-generation drug library and information resource. Nat. Med. 2017, 23, 405–408. [Google Scholar] [CrossRef] [Green Version]
- Hudson, T.J.; Anderson, W.; Artez, A.; Barker, A.D.; Bell, C.; Bernabe, R.R.; Bhan, M.K.; Calvo, F.; Eerola, I.; Gerhard, D.S.; et al. The International Cancer Genome Consortium International network of cancer genome projects. Nature 2010, 464, 993–998. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Levine, D.A. The Cancer Genome Atlas Research Network: Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Tsimberidou, A.; Wen, S.; Hong, D.S.; Wheler, J.J.; Falchook, G.S.; Fu, S.; Piha-Paul, S.; Naing, A.; Janku, F.; Aldape, K.; et al. Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: Validation and landmark analyses. Clin. Cancer Res. 2014, 20, 4827–4836. [Google Scholar] [CrossRef] [Green Version]
- André, F.; Bachelot, T.; Commo, F.; Campone, M.; Arnedos, M.; Dieras, V.; Lacroix-Triki, M.; Lacroix, L.; Cohen, P.; Gentien, D.; et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: A multicentre, prospective trial (SAFIR01/UNICANCER). Lancet Oncol. 2014, 15, 267–274. [Google Scholar] [CrossRef]
- Vansteenkiste, J.; Canon, J.-L.; De Braud, F.; Grossi, F.; De Pas, T.; Gray, J.E.; Su, W.-C.; Felip, E.; Yoshioka, H.; Gridelli, C.; et al. Safety and Efficacy of Buparlisib (BKM120) in Patients with PI3K Pathway-Activated Non-Small Cell Lung Cancer: Results from the Phase II BASALT-1 Study. J. Thorac. Oncol. 2015, 10, 1319–1327. [Google Scholar] [CrossRef] [Green Version]
- Garraway, L.A.; Sellers, W.R. Lineage dependency and lineage-survival oncogenes in human cancer. Nat. Rev. Cancer 2006, 6, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.V.; Haber, D.A.; Settleman, J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat. Rev. Cancer 2010, 10, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Goodspeed, A.; Heiser, L.M.; Gray, J.W.; Costello, J.C. Tumor-Derived Cell Lines as Molecular Models of Cancer Pharmacogenomics. Mol. Cancer Res. 2015, 14, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e16. [Google Scholar] [CrossRef] [Green Version]
- McMillan, E.A.; Ryu, M.-J.; Diep, C.H.; Mendiratta, S.; Clemenceau, J.R.; Vaden, R.M.; Kim, J.-H.; Motoyaji, T.; Covington, K.R.; Peyton, M.; et al. Chemistry-First Approach for Nomination of Personalized Treatment in Lung Cancer. Cell 2018, 173, 864–878.e29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-K.; Liu, Z.; Sa, J.K.; Shin, S.; Wang, J.; Bordyuh, M.; Cho, H.J.; Elliott, O.; Chu, T.; Choi, S.W.; et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat. Genet. 2018, 50, 1399–1411. [Google Scholar] [CrossRef]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Gonçalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef] [Green Version]
- Holbeck, S.L.; Collins, J.M.; Doroshow, J.H. Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol. Cancer Ther. 2010, 9, 1451–1460. [Google Scholar] [CrossRef] [Green Version]
- Garnett, M.J.; Edelman, E.J.; Heidorn, S.J.; Greenman, C.D.; Dastur, A.; Lau, K.W.; Greninger, P.; Thompson, I.R.; Luo, X.; Soares, J.; et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483, 570–575. [Google Scholar] [CrossRef] [Green Version]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschênes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.H.; et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, S.J.; Decker, B.; Roberts, E.; Horowitz, N.S.; Muto, M.G.; Worley, M.J.; Feltmate, C.M.; Nucci, M.R.; Swisher, E.M.; Nguyen, H.; et al. Prediction of DNA Repair Inhibitor Response in Short-Term Patient-Derived Ovarian Cancer Organoids. Cancer Discov. 2018, 8, 1404–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.-H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuveson, D.A.; Clevers, H. Cancer modeling meets human organoid technology. Science 2019, 364, 952–955. [Google Scholar] [CrossRef]
- Van De Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; Van Houdt, W.; Van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.H.N.; Siu, H.C.; Law, S.; Ho, S.L.; Yue, S.S.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Ma, S.K.Y.; Lam, K.O.; et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 2018, 23, 882–897.e11. [Google Scholar] [CrossRef] [Green Version]
- Seino, T.; Kawasaki, S.; Shimokawa, M.; Tamagawa, H.; Toshimitsu, K.; Fujii, M.; Ohta, Y.; Matano, M.; Nanki, K.; Kawasaki, K.; et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 2018, 22, 454–467.e6. [Google Scholar] [CrossRef] [Green Version]
- Sachs, N.; De Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373–386.e10. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Hu, W.; Matulay, J.T.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.-W.; Barlow, L.J.; Kandoth, C.; Williams, A.B.; et al. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 2018, 173, 515–528.e17. [Google Scholar] [CrossRef] [Green Version]
- Beshiri, M.L.; Tice, C.M.; Tran, C.; Nguyen, H.M.; Sowalsky, A.G.; Agarwal, S.; Jansson, K.H.; Yang, Q.; McGowen, K.M.; Yin, J.J.; et al. A PDX/Organoid Biobank of Advanced Prostate Cancers Captures Genomic and Phenotypic Heterogeneity for Disease Modeling and Therapeutic Screening. Clin. Cancer Res. 2018, 24, 4332–4345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calandrini, C.; Schutgens, F.; Oka, R.; Margaritis, T.; Candelli, T.; Mathijsen, L.; Ammerlaan, C.; Van Ineveld, R.L.; Derakhshan, S.; De Haan, S.; et al. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat. Commun. 2020, 11, 1310. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell 2020, 180, 188–204.e22. [Google Scholar] [CrossRef] [PubMed]
- Calles, A.; Rubio-Viqueira, B.; Hidalgo, M. Primary Human Non-Small Cell Lung and Pancreatic Tumorgraft Models-Utility and Applications in Drug Discovery and Tumor Biology. Curr. Protoc. Pharmacol. 2013, 61, 14.26.1–14.26.21. [Google Scholar] [CrossRef]
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumor xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 2012, 9, 338–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.; Caldas, C.; Clarke, R.; De Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef]
- Townsend, E.C.; Murakami, M.A.; Christodoulou, A.; Christie, A.L.; Köster, J.; DeSouza, T.A.; Morgan, E.A.; Kallgren, S.P.; Liu, H.; Wu, S.-C.; et al. The Public Repository of Xenografts (ProXe) enables discovery and randomized phase II-like trials in mice. Cancer Cell 2016, 29, 574–586. [Google Scholar] [CrossRef]
- Bruna, A.; Rueda, O.M.; Greenwood, W.; Batra, A.S.; Callari, M.; Batra, R.N.; Pogrebniak, K.; Sandoval, J.L.; Cassidy, J.W.; Vidakovic, A.T.; et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell 2016, 167, 260–274.e22. [Google Scholar] [CrossRef] [Green Version]
- Brait, M.; Izumchenko, E.; Kagohara, L.T.; Long, S.; Wysocki, P.T.; Faherty, B.; Fertig, E.; Khor, T.O.; Bruckheimer, E.; Baia, G.; et al. Comparative mutational landscape analysis of patient-derived tumour xenografts. Br. J. Cancer 2017, 116, 515–523. [Google Scholar] [CrossRef]
- Benini, S.; Gamberi, G.; Cocchi, S.; Garbetta, J.; Alberti, L.; Righi, A.; Gambarotti, M.; Picci, P.; Ferrari, S. Detection of circulating tumor cells in liquid biopsy from Ewing sarcoma patients. Cancer Manag. Res. 2018, 10, 49–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, M.; Zhu, P.; Mccarty, G.; Meyer, C.F.; Pratilas, C.A.; Levin, A.; Morris, C.D.; Albert, C.M.; Jackson, K.W.; Tang, C.-M.; et al. Size-based detection of sarcoma circulating tumor cells and cell clusters. Oncotarget 2017, 8, 78965–78977. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, H.H.; Neumann, H.A.; Henß, H.; Koch, H.; Kaiser, D.; Arnold, H. Development of Three Human Small Cell Lung Cancer Models in Nude Mice. Adv. Struct. Saf. Stud. 1985, 97, 77–86. [Google Scholar] [CrossRef]
- Ben-David, U.; Ha, G.; Tseng, Y.-Y.; Greenwald, N.F.; Oh, C.; Shih, J.; McFarland, J.M.; Wong, B.; Boehm, J.S.; Beroukhim, R.; et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 2017, 49, 1567–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-David, U.; Beroukhim, R.; Golub, T.R. Genomic evolution of cancer models: Perils and opportunities. Nat. Rev. Cancer 2018, 19, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Derose, Y.S.; Wang, G.; Lin, Y.-C.; Bernard, P.S.; Buys, S.S.; Ebbert, M.T.W.; Factor, R.; Matsen, C.B.; Milash, B.A.; Nelson, E.; et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011, 17, 1514–1520. [Google Scholar] [CrossRef]
- Cassidy, J.W.; Caldas, C.; Bruna, A. Maintaining Tumor Heterogeneity in Patient-Derived Tumor Xenografts. Cancer Res. 2015, 75, 2963–2968. [Google Scholar] [CrossRef] [Green Version]
- Hylander, B.L.; Punt, N.; Tang, H.; Hillman, J.; Vaughan, M.; Bshara, W.; Pitoniak, R.; Repasky, E.A. Origin of the vasculature supporting growth of primary patient tumor xenografts. J. Transl. Med. 2013, 11, 110. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Pirollo, K.F.; Tang, W.-H.; Rait, A.; Chang, E.H. Transferrin-Liposome-Mediated Systemic p53 Gene Therapy in Combination with Radiation Results in Regression of Human Head and Neck Cancer Xenografts. Hum. Gene Ther. 1999, 10, 2941–2952. [Google Scholar] [CrossRef]
- Sanz, L.; Cuesta, A.M.; Salas, C.; Corbacho, C.; Bellas, C.; Álvarez-Vallina, L. Differential transplantability of human endothelial cells in colorectal cancer and renal cell carcinoma primary xenografts. Lab. Investig. 2008, 89, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Boulay, G.; Sandoval, G.J.; Riggi, N.; Iyer, S.; Buisson, R.; Naigles, B.; Awad, M.E.; Rengarajan, S.; Volorio, A.; McBride, M.; et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171, 163–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erkizan, H.V.; Kong, Y.; Merchant, M.; Schlottmann, S.; Barber-Rotenberg, J.S.; Yuan, L.; Abaan, O.D.; Chou, T.-H.; Dakshanamurthy, S.; Brown, M.L.; et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing’s sarcoma. Nat. Med. 2009, 15, 750–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, T.; Surdez, D.; Rantala, J.K.; Haapa-Paananen, S.; Ban, J.; Kauer, M.; Tomazou, E.M.; Fey, V.; Alonso, J.; Kovar, H.; et al. High-throughput RNAi screen in Ewing sarcoma cells identifies leucine rich repeats and WD repeat domain containing 1 (LRWD1) as a regulator of EWS-FLI1 driven cell viability. Gene 2017, 596, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Brenner, J.C.; Feng, F.Y.; Han, S.; Patel, S.; Goyal, S.V.; Bou-Maroun, L.M.; Liu, M.; Lonigro, R.; Prensner, J.R.; Tomlins, S.A.; et al. PARP-1 inhibition as a targeted strategy to treat Ewing’s sarcoma. Cancer Res. 2012, 72, 1608–1613. [Google Scholar] [CrossRef] [Green Version]
- Stewart, E.; Goshorn, R.; Bradley, C.; Griffiths, L.M.; Benavente, C.A.; Twarog, N.R.; Miller, G.M.; Caufield, W.V.; Freeman, B.B.; Bahrami, A.; et al. Targeting the DNA repair pathway in Ewing sarcoma. Cell Rep. 2014, 9, 829–841. [Google Scholar] [CrossRef] [Green Version]
- Engert, F.; Schneider, C.; Weiss, L.M.; Probst, M.; Fulda, S. PARP Inhibitors Sensitize Ewing Sarcoma Cells to Temozolomide-Induced Apoptosis via the Mitochondrial Pathway. Mol. Cancer Ther. 2015, 14, 2818–2830. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-J.; Yoon, C.; Schmidt, B.; Park, J.; Zhang, A.Y.; Erkizan, H.V.; Toretsky, J.A.; Kirsch, D.G.; Yoon, S.S. Combining PARP-1 inhibition and radiation in Ewing sarcoma results in lethal DNA damage. Mol. Cancer Ther. 2013, 12, 2591–2600. [Google Scholar] [CrossRef] [Green Version]
- Choy, E.; Butrynski, J.E.; Harmon, D.C.; Morgan, J.A.; George, S.; Wagner, A.J.; D’Adamo, D.; Cote, G.M.; Flamand, Y.; Benes, C.H.; et al. Phase II study of olaparib in patients with refractory Ewing sarcoma following failure of standard chemotherapy. BMC Cancer 2014, 14, 813. [Google Scholar] [CrossRef] [Green Version]
- Franzetti, G.-A.; Laud-Duval, K.; Van Der Ent, W.; Brisac, A.; Irondelle, M.; Aubert, S.; Dirksen, U.; Bouvier, C.; De Pinieux, G.; Snaar-Jagalska, E.; et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene 2017, 36, 3505–3514. [Google Scholar] [CrossRef] [Green Version]
- Bairoch, A. The Cellosaurus, a Cell-Line Knowledge Resource. J. Biomol. Tech. JBT 2018, 29, 25. [Google Scholar] [CrossRef]
- Rubinstein, L.V.; Crowley, J.; Ivy, P.; Leblanc, M.; Sargent, D.J. Randomized phase II designs. Clin. Cancer Res. 2009, 15, 1883–1890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilding, J.L.; Bodmer, W.F. Cancer Cell Lines for Drug Discovery and Development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanni, P.; Landuzzi, L.; Manara, M.C.; Righi, A.; Nicoletti, G.; Cristalli, C.; Pasello, M.; Parra, A.; Carrabotta, M.; Ferracin, M.; et al. Bone sarcoma patient-derived xenografts are faithful and stable preclinical models for molecular and therapeutic investigations. Sci. Rep. 2019, 9, 12174. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M. Patient-derived orthotopic xenografts: Better mimic of metastasis than subcutaneous xenografts. Nat. Rev. Cancer 2015, 15, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Miyake, K.; Kiyuna, T.; Kawaguchi, K.; Higuchi, T.; Oshiro, H.; Zhang, Z.; Wangsiricharoen, S.; Razmjooei, S.; Li, Y.; Nelson, S.D.; et al. Regorafenib regressed a doxorubicin-resistant Ewing’s sarcoma in a patient-derived orthotopic xenograft (PDOX) nude mouse model. Cancer Chemother. Pharmacol. 2019, 83, 809–815. [Google Scholar] [CrossRef]
- Miyake, K.; Murakami, T.; Kiyuna, T.; Igarashi, K.; Kawaguchi, K.; Miyake, M.; Li, Y.; Nelson, S.D.; Dry, S.M.; Bouvet, M.; et al. The combination of temozolomide-irinotecan regresses a doxorubicin-resistant patient-derived orthotopic xenograft (PDOX) nude-mouse model of recurrent Ewing’s sarcoma with a FUS-ERG fusion and CDKN2A deletion: Direction for third-line patient therapy. Oncotarget 2017, 8, 103129–103136. [Google Scholar] [CrossRef] [Green Version]
- Murakami, T.; Singh, A.S.; Kiyuna, T.; Dry, S.M.; Li, Y.; James, A.W.; Igarashi, K.; Kawaguchi, K.; Delong, J.C.; Zhang, Y.; et al. Effective molecular targeting of CDK4/6 and IGF-1R in a rare FUS-ERG fusion CDKN2A-deletion doxorubicin-resistant Ewing’s sarcoma patient-derived orthotopic xenograft (PDOX) nude-mouse model. Oncotarget 2016, 7, 47556–47564. [Google Scholar] [CrossRef]
- Murakami, T.; Li, S.; Han, Q.; Tan, Y.; Kiyuna, T.; Igarashi, K.; Kawaguchi, K.; Hwang, H.K.; Miyake, K.; Singh, A.S.; et al. Recombinant methioninase effectively targets a Ewing’s sarcoma in a patient-derived orthotopic xenograft (PDOX) nude-mouse model. Oncotarget 2017, 8, 35630–35638. [Google Scholar] [CrossRef] [Green Version]
- Pawelek, J.M.; Low, K.B.; Bermudes, D. Bacteria as tumor-targeting vectors. Lancet Oncol. 2003, 4, 548–556. [Google Scholar] [CrossRef]
- Zhao, M.; Geller, J.; Ma, H.; Yang, M.; Penman, S.; Hoffman, R.M. Monotherapy with a tumor-targeting mutant of Salmonella typhimurium cures orthotopic metastatic mouse models of human prostate cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 10170–10174. [Google Scholar] [CrossRef] [Green Version]
- Miyake, K.; Kiyuna, T.; Li, S.; Han, Q.; Tan, Y.; Zhao, M.; Oshiro, H.; Kawaguchi, K.; Higuchi, T.; Zhang, Z.; et al. Combining Tumor-Selective Bacterial Therapy with Salmonella typhimurium A1-R and Cancer Metabolism Targeting with Oral Recombinant Methioninase Regressed an Ewing’s Sarcoma in a Patient-Derived Orthotopic Xenograft Model. Chemotherapy 2018, 63, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Stebbing, J.; Paz, K.; Schwartz, G.K.; Wexler, L.H.; Maki, R.G.; Pollock, R.E.; Morris, R.; Cohen, R.; Shankar, A.; Blackman, G.; et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer 2014, 120, 2006–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondo, T. Current Status and Perspectives of Patient-Derived Models for Ewing’s Sarcoma. Cancers 2020, 12, 2520. https://doi.org/10.3390/cancers12092520
Kondo T. Current Status and Perspectives of Patient-Derived Models for Ewing’s Sarcoma. Cancers. 2020; 12(9):2520. https://doi.org/10.3390/cancers12092520
Chicago/Turabian StyleKondo, Tadashi. 2020. "Current Status and Perspectives of Patient-Derived Models for Ewing’s Sarcoma" Cancers 12, no. 9: 2520. https://doi.org/10.3390/cancers12092520