Optimized 3D Bioprinting Technology Based on Machine Learning: A Review of Recent Trends and Advances
Abstract
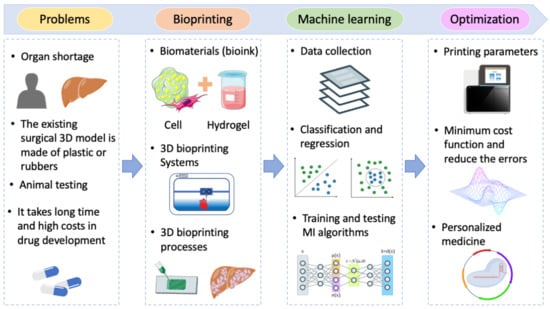
:1. Introduction
2. Machine-Learning Principles Used in 3D Bioprinting
2.1. Materials and Modalities of 3D Bioprinting
2.2. Machine Learning
2.3. Bioprinting Process with Machine Learning
3. Challenges and Future Prospects
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mao, A.S.; Mooney, D.J. Regenerative medicine: Current therapies and future directions. Proc. Natl. Acad. Sci. USA 2015, 112, 14452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skardal, A.; Shupe, T.; Atala, A. Body-on-a-Chip: Regenerative Medicine for Personalized Medicine. Princ. Regen. Med. 2019, 769–786. [Google Scholar]
- Risse, G.B.; Warner, J.H. Reconstructing clinical activities: Patient records in medical history. Soc. Hist. Med. 1992, 5, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Hamburg, M.A.; Collins, F.S. The path to personalized medicine. N. Engl. J. Med. 2010, 363, 301–304. [Google Scholar] [CrossRef]
- Jovic, T.H.; Combellack, E.J.; Jessop, Z.M.; Whitaker, I.S. 3D Bioprinting and the Future of Surgery. Front. Surg. 2020, 129. [Google Scholar] [CrossRef]
- Mironov, V.; Kasyanov, V.; Markwald, R.R. Organ printing: From bioprinter to organ biofabrication line. Curr. Opin. Biotechnol. 2011, 22, 667–673. [Google Scholar] [CrossRef]
- Ramesh, S.; Harrysson, O.L.A.; Rao, P.K.; Tamayol, A.; Cormier, D.R.; Zhang, Y.; Rivero, I.V. Extrusion bioprinting: Recent progress, challenges, and future opportunities. Bioprinting 2021, 21, e00116. [Google Scholar] [CrossRef]
- Li, X.; Liu, B.; Pei, B.; Chen, J.; Zhou, D.; Peng, J.; Zhang, X.; Jia, W.; Xu, T. Inkjet Bioprinting of Biomaterials. Chem. Rev. 2020, 120, 10793–10833. [Google Scholar] [CrossRef]
- Wang, Z.; Abdulla, R.; Parker, B.; Samanipour, R.; Ghosh, S.; Kim, K. A simple and high-resolution stereolithography-based 3D bioprinting system using visible light crosslinkable bioinks. Biofabrication 2015, 7, 045009. [Google Scholar] [CrossRef]
- Nair, K.; Gandhi, M.; Khalil, S.; Yan, K.C.; Marcolongo, M.; Barbee, K.; Sun, W. Characterization of cell viability during bioprinting processes. Biotechnol. J. 2009, 4, 1168–1177. [Google Scholar] [CrossRef]
- Delli, U.; Chang, S. Automated Process Monitoring in 3D Printing Using Supervised Machine Learning. Procedia Manuf. 2018, 26, 865–870. [Google Scholar] [CrossRef]
- Ji, C.; Mandania, R.; Liu, J.; Liret, A.; Kern, M. Incorporating Risk in Field Services Operational Planning Process. In International Conference on Innovative Techniques and Applications of Artificial Intelligence; Springer: Cham, Switzerland; Cambridge, UK, 2018; Volume 11311 LNAI, ISBN 9783030041908. [Google Scholar]
- Goh, G.D.; Sing, S.L.; Yeong, W.Y. A review on machine learning in 3D printing: Applications, potential, and challenges. Artif. Intell. Rev. 2021, 54, 63–94. [Google Scholar] [CrossRef]
- Menon, A.; Póczos, B.; Feinberg, A.W.; Washburn, N.R. Optimization of Silicone 3D Printing with Hierarchical Machine Learning. 3D Print. Addit. Manuf. 2019, 6, 181–189. [Google Scholar] [CrossRef]
- Ruberu, K.; Senadeera, M.; Rana, S.; Gupta, S.; Chung, J.; Yue, Z.; Venkatesh, S.; Wallace, G. Coupling machine learning with 3D bioprinting to fast track optimisation of extrusion printing. Appl. Mater. Today 2021, 22, 100914. [Google Scholar] [CrossRef]
- Visconti, R.P.; Kasyanov, V.; Gentile, C.; Zhang, J.; Markwald, R.R.; Mironov, V. Towards organ printing: Engineering an intra-organ branched vascular tree. Expert Opin. Biol. Ther. 2010, 10, 409–420. [Google Scholar] [CrossRef]
- Fedorovich, N.E.; Alblas, J.; De Wijn, J.R.; Hennink, W.E.; Verbout, A.B.J.; Dhert, W.J.A. Hydrogels as extracellular matrices for skeletal tissue engineering: State-of-the-art and novel application in organ printing. Tissue Eng. 2007, 13, 1905–1925. [Google Scholar] [CrossRef]
- Mironov, V.; Boland, T.; Trusk, T.; Forgacs, G.; Markwald, R.R. Organ printing: Computer-aided jet-based 3D tissue engineering. Trends Biotechnol. 2003, 21, 157–161. [Google Scholar] [CrossRef]
- Gopinathan, J.; Noh, I. Recent trends in bioinks for 3D printing. Biomater. Res. 2018, 22, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Park, M.N.; Kim, J.; Jang, J.; Kim, H.K.; Cho, D.W. Characterization of cornea-specific bioink: High transparency, improved in vivo safety. J. Tissue Eng. 2019, 10, 2041731418823382. [Google Scholar] [CrossRef] [Green Version]
- Rutz, A.L.; Hyland, K.E.; Jakus, A.E.; Burghardt, W.R.; Shah, R.N. A Multimaterial Bioink Method for 3D Printing Tunable, Cell-Compatible Hydrogels. Adv. Mater. 2015, 27, 1607–1614. [Google Scholar] [CrossRef] [Green Version]
- Tasoglu, S.; Demirci, U. Bioprinting for stem cell research. Trends Biotechnol. 2013, 31, 10–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reimann, F.; Habib, A.M.; Tolhurst, G.; Parker, H.E.; Rogers, G.J.; Gribble, F.M. Glucose Sensing in L Cells: A Primary Cell Study. Cell Metab. 2008, 8, 532–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandrycky, C.; Wang, Z.; Kim, K.; Kim, D.H. 3D bioprinting for engineering complex tissues. Biotechnol. Adv. 2016, 34, 422–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, A.S. Hydrogels for biomedical applications. Adv. Drug Deliv. Rev. 2012, 64, 18–23. [Google Scholar] [CrossRef]
- Li, J.; Wu, C.; Chu, P.K.; Gelinsky, M. 3D printing of hydrogels: Rational design strategies and emerging biomedical applications. Mater. Sci. Eng. R Rep. 2020, 140, 100543. [Google Scholar] [CrossRef]
- Skardal, A.; Atala, A. Biomaterials for Integration with 3-D Bioprinting. Ann. Biomed. Eng. 2015, 43, 730–746. [Google Scholar] [CrossRef] [PubMed]
- Mei, Q.; Rao, J.; Bei, H.P.; Liu, Y.; Zhao, X. 3D Bioprinting Photo-Crosslinkable Hydrogels for Bone and Cartilage Repair. Int. J. Bioprinting 2021, 7, 367. [Google Scholar] [CrossRef]
- Ozbolat, I.T.; Hospodiuk, M. Current advances and future perspectives in extrusion-based bioprinting. Biomaterials 2016, 76, 321–343. [Google Scholar] [CrossRef] [Green Version]
- Norotte, C.; Marga, F.S.; Niklason, L.E.; Forgacs, G. Scaffold-free vascular tissue engineering using bioprinting. Biomaterials 2009, 30, 5910–5917. [Google Scholar] [CrossRef] [Green Version]
- Duocastella, M.; Colina, M.; Fernández-Pradas, J.M.; Serra, P.; Morenza, J.L. Study of the laser-induced forward transfer of liquids for laser bioprinting. Appl. Surf. Sci. 2007, 253, 7855–7859. [Google Scholar] [CrossRef]
- Peng, W.; Datta, P.; Ayan, B.; Ozbolat, V.; Sosnoski, D.; Ozbolat, I.T. 3D bioprinting for drug discovery and development in pharmaceutics. Acta Biomater. 2017, 57, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Kirchmajer, D.M.; Gorkin, R., III; in het Panhuis, M. An overview of the suitability of hydrogel-forming polymers for extrusion-based 3D-printing. J. Mater. Chem. B 2015, 3, 4105–4117. [Google Scholar] [CrossRef] [PubMed]
- Pescosolido, L.; Schuurman, W.; Malda, J.; Matricardi, P.; Alhaique, F.; Coviello, T.; Van Weeren, P.R.; Dhert, W.J.A.; Hennink, W.E.; Vermonden, T. Hyaluronic acid and dextran-based semi-IPN hydrogels as biomaterials for bioprinting. Biomacromolecules 2011, 12, 1831–1838. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.W.; Lee, J.S.; Cho, D.W. Computer-Aided multiple-head 3D printing system for printing of heterogeneous organ/tissue constructs. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Alghuwainem, A.; Alshareeda, A.T.; Alsowayan, B. Scaffold-free 3-D cell sheet technique bridges the gap between 2-D cell culture and animal models. Int. J. Mol. Sci. 2019, 20, 4926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serra, P.; Piqué, A. Laser-Induced Forward Transfer: Fundamentals and Applications. Adv. Mater. Technol. 2019, 4, 1–33. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Chang, Y.K.; Heung, B.; Nguyen-Quang, T.; Price, G.W.; Al-Mallahi, A. A deep learning approach for RGB image-based powdery mildew disease detection on strawberry leaves. Comput. Electron. Agric. 2021, 183, 106042. [Google Scholar] [CrossRef]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Joachims, T. Transductive Learning via Spectral Graph Partitioning. Proc. Twent. Int. Conf. Mach. Learn. 2003, 1, 290–297. [Google Scholar]
- Singh, A.; Thakur, N.; Sharma, A. A review of supervised machine learning algorithms. In Proceedings of the 2016 3rd International Conference on Computing for Sustainable Global Development (INDIACom), New Delhi, India, 16–18 March 2016; pp. 1310–1315. [Google Scholar]
- Nasteski, V. An overview of the supervised machine learning methods. Horizons. B 2017, 4, 51–62. [Google Scholar] [CrossRef]
- Wu, M.; Phoha, V.V.; Moon, Y.B.; Belman, A.K. Detecting malicious defects in 3D printing process using machine learning and image classification. ASME Int. Mech. Eng. Congr. Expo. Proc. 2016, 14, 4–9. [Google Scholar]
- Wu, H.C.; Chen, T.C.T. Quality control issues in 3D-printing manufacturing: A review. Rapid Prototyp. J. 2018, 24, 607–614. [Google Scholar] [CrossRef]
- Gu, G.X.; Chen, C.T.; Buehler, M.J. De novo composite design based on machine learning algorithm. Extrem. Mech. Lett. 2018, 18, 19–28. [Google Scholar] [CrossRef]
- Vosniakos, G.C.; Maroulis, T.; Pantelis, D. A method for optimizing process parameters in layer-based rapid prototyping. Proc. Inst. Mech. Eng. Part B J. Eng. Manuf. 2007, 221, 1329–1340. [Google Scholar] [CrossRef]
- Koeppe, A.; Hernandez Padilla, C.A.; Voshage, M.; Schleifenbaum, J.H.; Markert, B. Efficient numerical modeling of 3D-printed lattice-cell structures using neural networks. Manuf. Lett. 2018, 15, 147–150. [Google Scholar] [CrossRef]
- Asadi-Eydivand, M.; Solati-Hashjin, M.; Fathi, A.; Padashi, M.; Abu Osman, N.A. Optimal design of a 3D-printed scaffold using intelligent evolutionary algorithms. Appl. Soft Comput. J. 2016, 39, 36–47. [Google Scholar] [CrossRef]
- Vahabli, E.; Rahmati, S. Application of an RBF neural network for FDM parts’ surface roughness prediction for enhancing surface quality. Int. J. Precis. Eng. Manuf. 2016, 17, 1589–1603. [Google Scholar] [CrossRef]
- He, H.; Yang, Y.; Pan, Y. Machine learning for continuous liquid interface production: Printing speed modelling. J. Manuf. Syst. 2019, 50, 236–246. [Google Scholar] [CrossRef]
- Gobert, C.; Reutzel, E.W.; Petrich, J.; Nassar, A.R.; Phoha, S. Application of supervised machine learning for defect detection during metallic powder bed fusion additive manufacturing using high resolution imaging. Addit. Manuf. 2018, 21, 517–528. [Google Scholar] [CrossRef]
- Olaode, A.; Naghdy, G.; Todd, C. Unsupervised Classification of Images: A Review. Int. J. Image Process. 2014, 8, 325. [Google Scholar]
- Längkvist, M.; Karlsson, L.; Loutfi, A. A review of unsupervised feature learning and deep learning for time-series modeling. Pattern Recognit. Lett. 2014, 42, 11–24. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.; Jiang, J. A perspective on Using Machine Learning in 3D Bioprinting. Int. J. Bioprinting 2020, 6, 4–11. [Google Scholar] [CrossRef]
- Zhu, X.; Goldberg, A.B. Introduction to Semi-Supervised Learning. In Synthesis Lectures on Artificial Intelligence and Machine Learning; Morgan & Claypool Publishers: Williston, ND, USA, 2009; Volume 6, ISBN 9781598295481. [Google Scholar]
- Van Engelen, J.E.; Hoos, H.H. A survey on semi-supervised learning. Mach. Learn. 2020, 109, 373–440. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, S.; Zhou, Z.H. New semi-supervised classification method based on modified cluster assumption. IEEE Trans. Neural Netw. Learn. Syst. 2012, 23, 689–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Wang, S. Semi-supervised learning via regularized boosting working on multiple semi-supervised assumptions. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, C.; Hebert, M.; Schneiderman, H. Semi-supervised self-training of object detection models. Proc.-Seventh IEEE Work. Appl. Comput. Vision, WACV; 2005; pp. 29–36. [Google Scholar]
- Luo, Y.; Zhu, J.; Li, M.; Ren, Y.; Zhang, B. Smooth neighbors on teacher graphs for semi-supervised learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8896–8905. [Google Scholar]
- Wiering, M.; Otterlo, M.V. Conclusions, Future Directions and Outlook. In Reinforcement Learning; Springer: Berlin/Heidelberg, Germany, 2012; pp. 613–630. [Google Scholar]
- Wang, H.N.; Liu, N.; Zhang, Y.Y.; Feng, D.W.; Huang, F.; Li, D.S.; Zhang, Y. ming Deep reinforcement learning: A survey. Front. Inf. Technol. Electron. Eng. 2020, 21, 1726–1744. [Google Scholar] [CrossRef]
- Guan, J.; You, S.; Xiang, Y.; Schimelman, J.; Alido, J.; Ma, X.; Tang, M.; Chen, S. Compensating the cell-induced light scattering effect in light-based bioprinting using deep learning. Biofabrication 2022, 14, 015011. [Google Scholar] [CrossRef]
- Datta, P.; Barui, A.; Wu, Y.; Ozbolat, V.; Moncal, K.K.; Ozbolat, I.T. Essential steps in bioprinting: From pre- to post-bioprinting. Biotechnol. Adv. 2018, 36, 1481–1504. [Google Scholar] [CrossRef] [PubMed]
- Passamai, V.E.; Dernowsek, J.A.; Nogueira, J.; Lara, V.; Vilalba, F.; Mironov, V.A.; Rezende, R.A.; Da Silva, J.V. From 3D Bioprinters to a fully integrated Organ Biofabrication Line. J. Phys. Conf. Ser. 2016, 705, 012010. [Google Scholar] [CrossRef]
- Lee, J.; Oh, S.J.; An, S.H.; Kim, W.-D.; Kim, S.-H. Machine learning-based design strategy for 3D printable bioink: Elastic modulus and yield stress determine printability. Biofabrication 2020, 12, 035018. [Google Scholar] [CrossRef]
- Shi, J.; Song, J.; Song, B.; Lu, W.F. Multi-Objective Optimization Design through Machine Learning for Drop-on-Demand Bioprinting. Engineering 2019, 5, 586–593. [Google Scholar] [CrossRef]
- Ng, W.L.; Chan, A.; Ong, Y.S.; Chua, C.K. Deep learning for fabrication and maturation of 3D bioprinted tissues and organs. Virtual Phys. Prototyp. 2020, 15, 340–358. [Google Scholar] [CrossRef]
- Yuk, H.; Zhao, X. A New 3D Printing Strategy by Harnessing Deformation, Instability, and Fracture of Viscoelastic Inks. Adv. Mater. 2018, 30, 1704028. [Google Scholar] [CrossRef]
- Scime, L.; Beuth, J. Using machine learning to identify in-situ melt pool signatures indicative of flaw formation in a laser powder bed fusion additive manufacturing process. Addit. Manuf. 2019, 25, 151–165. [Google Scholar] [CrossRef]
- Caggiano, A.; Zhang, J.; Alfieri, V.; Caiazzo, F.; Gao, R.; Teti, R. Machine learning-based image processing for on-line defect recognition in additive manufacturing. CIRP Ann. 2019, 68, 451–454. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, S.; Shin, Y.C. In-Process monitoring of porosity during laser additive manufacturing process. Addit. Manuf. 2019, 28, 497–505. [Google Scholar] [CrossRef]
- Pepper, M.E.; Cass, C.A.P.; Mattimore, J.P.; Burg, T.; Booth, B.W.; Burg, K.J.L.; Groff, R.E. Post-bioprinting processing methods to improve cell viability and pattern fidelity in heterogeneous tissue test systems. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 259–262. [Google Scholar]
- Zhao, H.; Xu, J.; Zhang, E.; Qi, R.; Huang, Y.; Lv, F.; Liu, L.; Gu, Q.; Wang, S. 3D Bioprinting of Polythiophene Materials for Promoting Stem Cell Proliferation in a Nutritionally Deficient Environment. ACS Appl. Mater. Interfaces 2021, 13, 25759–25770. [Google Scholar] [CrossRef]
- Cubo, N.; Garcia, M.; Del Cañizo, J.F.; Velasco, D.; Jorcano, J.L. 3D bioprinting of functional human skin: Production and in vivo analysis. Biofabrication 2017, 9, 015006. [Google Scholar] [CrossRef] [Green Version]
- Conev, A.; Litsa, E.E.; Perez, M.R.; Diba, M.; Mikos, A.G.; Kavraki, L.E. Machine learning-guided three-dimensional printing of tissue engineering scaffolds. Tissue Eng.-Part A 2020, 26, 1359–1368. [Google Scholar] [CrossRef]
- Jin, Z.; Zhang, Z.; Shao, X.; Gu, G.X. Monitoring Anomalies in 3D Bioprinting with Deep Neural Networks. ACS Biomater. Sci. Eng. 2021. [Google Scholar] [CrossRef]
- An, J.; Chua, C.K.; Mironov, V. Application of Machine Learning in 3D Bioprinting: Focus on Development of Big Data and Digital Twin. Int. J. Bioprinting 2021, 7, 342. [Google Scholar] [CrossRef] [PubMed]
- Maceachern, S.J.; Forkert, N.D. Machine learning for precision medicine. Genome 2021, 64, 416–425. [Google Scholar] [CrossRef] [PubMed]






| Algorithm | References |
|---|---|
| Naive bayes | [43] |
| Decision tree | [44] |
| Convolutional neural network | [45] |
| Genetic programming | [46] |
| Long short-term memory | [47] |
| Particle swarm algorithm | [48] |
| K-nearest neighbour | [43] |
| Radial basis function | [49] |
| Siamese neural network | [50] |
| Support vector machine | [51] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.; Lee, Y.; Li, Z.; Hu, J.; Park, S.S.; Kim, K. Optimized 3D Bioprinting Technology Based on Machine Learning: A Review of Recent Trends and Advances. Micromachines 2022, 13, 363. https://doi.org/10.3390/mi13030363
Shin J, Lee Y, Li Z, Hu J, Park SS, Kim K. Optimized 3D Bioprinting Technology Based on Machine Learning: A Review of Recent Trends and Advances. Micromachines. 2022; 13(3):363. https://doi.org/10.3390/mi13030363
Chicago/Turabian StyleShin, Jaemyung, Yoonjung Lee, Zhangkang Li, Jinguang Hu, Simon S. Park, and Keekyoung Kim. 2022. "Optimized 3D Bioprinting Technology Based on Machine Learning: A Review of Recent Trends and Advances" Micromachines 13, no. 3: 363. https://doi.org/10.3390/mi13030363