Ribotoxic Proteins, Known as Inhibitors of Protein Synthesis, from Mushrooms and Other Fungi According to Endo’s Fragment Detection
Abstract
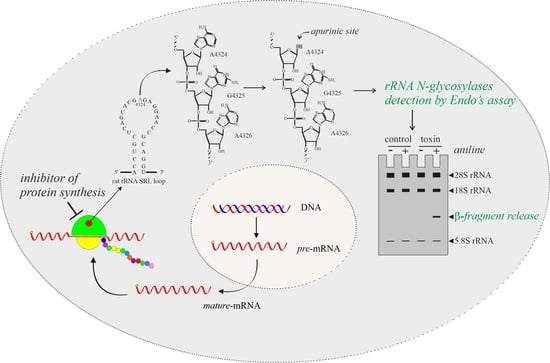
:1. Introduction
2. Fungal RIPs Able to Release the β-Fragment
2.1. Calcaelin from Calvatia caelata
2.2. Lyophyllin from Lyophyllum shimeji
2.3. Marmorin from Hypsizygus marmoreus
2.4. Mucoricin from Rhizopus delemar
2.5. Volvarin from Volvariella valvacea
3. Other Ribotoxic Enzymes from Fungi Improperly Classified as N-Glycosylases
3.1. Bolesatine from Boletus satanas
3.2. Flammin from Flammulina velutipes
3.3. Flammulin from Flammulina velutipes
3.4. Hypsin from Hypsizygus marmoreus
3.5. Pleuturegin from Pleurotus tuber-regium
3.6. Tricholin from Trichoderma viride
3.7. Velin from Flammulina velutipes
3.8. Velutin from Flammulina velutipes
3.9. Mushroom Type 2 Ribosome Inactivating Protein-like Genes
4. Isolation and Purification of Protein Synthesis Inhibitors from Mushrooms and Other Fungi
5. Sequence Alignment and Phylogenetic Analysis
6. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MEPs | peptidyl-Lys metalloendopeptidases |
| MS | mass spectrometry |
| RIPs | ribosome inactivating proteins |
| RL-Ps | ribotoxin-like proteins |
| SRL | sarcin ricin loop |
References
- Endo, Y.; Huber, P.W.; Wool, I.G. The ribonuclease activity of the cytotoxin alpha-sarcin. The characteristics of the enzymatic activity of alpha-sarcin with ribosomes and ribonucleic acids as substrates. J. Biol. Chem. 1983, 258, 2662–2667. [Google Scholar] [CrossRef]
- Grela, P.; Szajwaj, M.; Horbowicz-Drożdżal, P.; Tchórzewski, M. How Ricin Damages the Ribosome. Toxins 2019, 11, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olombrada, M.; Peña, C.; Rodríguez-Galán, O.; Klingauf-Nerurkar, P.; Portugal-Calisto, D.; Oborská-Oplová, M.; Altvater, M.; Gavilanes, J.G.; Martínez-Del-Pozo, Á.; de la Cruz, J.; et al. The ribotoxin α-sarcin can cleave the sarcin/ricin loop on late 60S pre-ribosomes. Nucleic Acids Res. 2020, 48, 6210–6222. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.K.; Wong, E.C.; Lee, K.M.; Wong, K.B. Structures of eukaryotic ribosomal stalk proteins and its complex with trichosanthin, and their implications in recruiting ribosome-inactivating proteins to the ribosomes. Toxins 2015, 7, 638–647. [Google Scholar] [CrossRef] [Green Version]
- Diaconu, M.; Kothe, U.; Schlünzen, F.; Fischer, N.; Harms, J.M.; Tonevitsky, A.G.; Stark, H.; Rodnina, M.V.; Wahl, M.C. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 2005, 121, 991–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohun, E.; Twardowski, T. alpha-Sarcin domain is a fragment of 23S and 26S rRNA strategic for ribosome function. Acta Biochim. Pol. 1993, 40, 12–16. [Google Scholar] [CrossRef] [Green Version]
- Herrero-Galán, E.; García-Ortega, L.; Olombrada, M.; Lacadena, J.; Del Pozo Á, M.; Gavilanes, J.G.; Oñaderra, M. Hirsutellin A: A Paradigmatic Example of the Insecticidal Function of Fungal Ribotoxins. Insects 2013, 4, 339–356. [Google Scholar] [CrossRef] [Green Version]
- Landi, N.; Pacifico, S.; Ragucci, S.; Iglesias, R.; Piccolella, S.; Amici, A.; Di Giuseppe, A.M.A.; Di Maro, A. Purification, characterization and cytotoxicity assessment of Ageritin: The first ribotoxin from the basidiomycete mushroom Agrocybe aegerita. Biochim. Biophys Acta Gen. Subj. 2017, 1861, 1113–1121. [Google Scholar] [CrossRef]
- Ragucci, S.; Landi, N.; Russo, R.; Valletta, M.; Pedone, P.V.; Chambery, A.; Di Maro, A. Ageritin from Pioppino Mushroom: The Prototype of Ribotoxin-Like Proteins, a Novel Family of Specific Ribonucleases in Edible Mushrooms. Toxins 2021, 13, 263. [Google Scholar] [CrossRef]
- Landi, N.; Ragucci, S.; Russo, R.; Valletta, M.; Pizzo, E.; Ferreras, J.M.; Di Maro, A. The ribotoxin-like protein Ostreatin from Pleurotus ostreatus fruiting bodies: Confirmation of a novel ribonuclease family expressed in basidiomycetes. Int. J. Biol. Macromol. 2020, 161, 1329–1336. [Google Scholar] [CrossRef]
- Olombrada, M.; Lázaro-Gorines, R.; López-Rodríguez, J.C.; Martínez-Del-Pozo, Á.; Oñaderra, M.; Maestro-López, M.; Lacadena, J.; Gavilanes, J.G.; García-Ortega, L. Fungal Ribotoxins: A Review of Potential Biotechnological Applications. Toxins 2017, 9, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landi, N.; Ragucci, S.; Culurciello, R.; Russo, R.; Valletta, M.; Pedone, P.V.; Pizzo, E.; Di Maro, A. Ribotoxin-like proteins from Boletus edulis: Structural properties, cytotoxicity and in vitro digestibility. Food Chem. 2021, 359, 129931. [Google Scholar] [CrossRef] [PubMed]
- Di Maro, A.; Citores, L.; Russo, R.; Iglesias, R.; Ferreras, J.M. Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome-inactivating proteins from angiosperms. Plant Mol. Biol. 2014, 85, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Landi, N.; Ruocco, M.R.; Ragucci, S.; Aliotta, F.; Nasso, R.; Pedone, P.V.; Di Maro, A. Quinoa as source of type 1 ribosome inactivating proteins: A novel knowledge for a revision of its consumption. Food Chem. 2021, 342, 128337. [Google Scholar] [CrossRef]
- Stirpe, F.; Gilabert-Oriol, R. Ribosome-Inactivating Proteins: An Overview. In Plant Toxins; Carlini, C.R., Ligabue-Braun, R., Gopalakrishnakone, P., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 153–182. [Google Scholar]
- Liu, R.S.; Yang, J.H.; Liu, W.Y. Isolation and enzymatic characterization of lamjapin, the first ribosome-inactivating protein from cryptogamic algal plant (Laminaria japonica A). Eur. J. Biochem. 2002, 269, 4746–4752. [Google Scholar] [CrossRef]
- O’Loughlin, E.V.; Robins-Browne, R.M. Effect of Shiga toxin and Shiga-like toxins on eukaryotic cells. Microbes Infect. 2001, 3, 493–507. [Google Scholar] [CrossRef]
- Pizzo, E.; Di Maro, A. A new age for biomedical applications of Ribosome Inactivating Proteins (RIPs): From bioconjugate to nanoconstructs. J. Biomed. Sci. 2016, 23, 54. [Google Scholar] [CrossRef] [Green Version]
- Ng, T.B. Peptides and proteins from fungi. Peptides 2004, 25, 1055–1073. [Google Scholar] [CrossRef]
- Stirpe, F. Ribosome-inactivating proteins. Toxicon 2004, 44, 371–383. [Google Scholar] [CrossRef]
- Schrot, J.; Weng, A.; Melzig, M.F. Ribosome-inactivating and related proteins. Toxins 2015, 7, 1556–1615. [Google Scholar] [CrossRef]
- Peumans, W.J.; Hao, Q.; Van Damme, E.J. Ribosome-inactivating proteins from plants: More than RNA N-glycosidases? FASEB J. 2001, 15, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Valbonesi, P.; Bonora, E.; Gorini, P.; Bolognesi, A.; Stirpe, F. Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: Effect on DNA, RNA and poly(A). Nucleic Acids Res. 1997, 25, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Aceto, S.; Di Maro, A.; Conforto, B.; Siniscalco, G.G.; Parente, A.; Delli Bovi, P.; Gaudio, L. Nicking activity on pBR322 DNA of ribosome inactivating proteins from Phytolacca dioica L. leaves. Biol. Chem. 2005, 386, 307–317. [Google Scholar] [CrossRef]
- Lapadula, W.J.; Sánchez Puerta, M.V.; Juri Ayub, M. Revising the taxonomic distribution, origin and evolution of ribosome inactivating protein genes. PLoS ONE 2013, 8, e72825. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Zhou, Y.K.; Ji, Z.L.; Chen, X.R. The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Front. Plant. Sci. 2018, 9, 146. [Google Scholar] [CrossRef] [Green Version]
- Rajamohan, F.; Pugmire, M.J.; Kurinov, I.V.; Uckun, F.M. Modeling and alanine scanning mutagenesis studies of recombinant pokeweed antiviral protein. J. Biol. Chem. 2000, 275, 3382–3390. [Google Scholar] [CrossRef] [Green Version]
- Endo, Y.; Tsurugi, K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J. Biol. Chem. 1988, 263, 8735–8739. [Google Scholar] [CrossRef]
- Olsnes, S.; Pihl, A. Ricin—A potent inhibitor of protein synthesis. FEBS Lett. 1972, 20, 327–329. [Google Scholar] [CrossRef] [Green Version]
- Kretz, O.; Creppy, E.E.; Dirheimer, G. Characterization of bolesatine, a toxic protein from the mushroom Boletus satanas Lenz and it’s effects on kidney cells. Toxicology 1991, 66, 213–224. [Google Scholar] [CrossRef]
- Kretz, O.; Creppy, E.E.; Dirheimer, G. Disposition of the toxic protein, bolesatine, in rats: Its resistance to proteolytic enzymes. Xenobiotica 1991, 21, 65–73. [Google Scholar] [CrossRef]
- Kretz, O.; Barbieri, L.; Creppy, E.E.; Dirheimer, G. Inhibition of protein synthesis in liver and kidney of mice by bolesatine: Mechanistic approaches to the mode of action at the molecular level. Toxicology 1992, 73, 297–304. [Google Scholar] [CrossRef]
- Ennamany, R.; Lavergne, J.P.; Reboud, J.P.; Dirheimer, G.; Creppy, E.E. Mode of action of bolesatine, a cytotoxic glycoprotein from Boletus satanas Lenz. Mechanistic approaches. Toxicology 1995, 100, 51–55. [Google Scholar] [CrossRef]
- Ng, T.B.; Lam, Y.W.; Wang, H. Calcaelin, a new protein with translation-inhibiting, antiproliferative and antimitogenic activities from the mosaic puffball mushroom Calvatia caelata. Planta Med. 2003, 69, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Q.; Shi, W.W.; Xiao, M.J.; Tang, Y.S.; Zheng, Y.T.; Shaw, P.C. Lyophyllin, a Mushroom Protein from the Peptidase M35 Superfamily Is an RNA N-Glycosidase. Int. J. Mol. Sci. 2021, 22, 11598. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.K.; Ng, T.B. First simultaneous isolation of a ribosome inactivating protein and an antifungal protein from a mushroom (Lyophyllum shimeji) together with evidence for synergism of their antifungal effects. Arch. Biochem. Biophys. 2001, 393, 271–280. [Google Scholar] [CrossRef]
- Wong, J.H.; Wang, H.X.; Ng, T.B. Marmorin, a new ribosome inactivating protein with antiproliferative and HIV-1 reverse transcriptase inhibitory activities from the mushroom Hypsizigus marmoreus. Appl. Microbiol. Biotechnol. 2008, 81, 669–674. [Google Scholar] [CrossRef]
- Soliman, S.S.M.; Baldin, C.; Gu, Y.; Singh, S.; Gebremariam, T.; Swidergall, M.; Alqarihi, A.; Youssef, E.G.; Alkhazraji, S.; Pikoulas, A.; et al. Mucoricin is a ricin-like toxin that is critical for the pathogenesis of mucormycosis. Nat. Microbiol. 2021, 6, 313–326. [Google Scholar] [CrossRef]
- Yao, Q.Z.; Yu, M.M.; Ooi, L.S.; Ng, T.B.; Chang, S.T.; Sun, S.S.; Ooi, V.E. Isolation and Characterization of a Type 1 Ribosome-Inactivating Protein from Fruiting Bodies of the Edible Mushroom (Volvariella volvacea). J. Agric. Food Chem. 1998, 46, 788–792. [Google Scholar] [CrossRef]
- Landi, N.; Clemente, A.; Pedone, P.V.; Ragucci, S.; Di Maro, A. An Updated Review of Bioactive Peptides from Mushrooms in a Well-Defined Molecular Weight Range. Toxins 2022, 14, 84. [Google Scholar] [CrossRef]
- Barbieri, L.; Battelli, M.G.; Stirpe, F. Ribosome-inactivating proteins from plants. Biochim. Biophys. Acta 1993, 1154, 237–282. [Google Scholar] [CrossRef]
- Kretz, O.; Reinbolt, J.; Creppy, E.E.; Dirheimer, G. Properties of bolesatine, a translational inhibitor from Boletus satanas Lenz. Amino-terminal sequence determination and inhibition of rat mitochondrial protein synthesis. Toxicol. Lett. 1992, 64–65, 763–766. [Google Scholar] [CrossRef]
- Kretz, O.; Creppy, E.E.; Boulanger, Y.; Dirheimer, G. Purification and some properties of bolesatine, a protein inhibiting in vitro protein synthesis, from the mushroom Boletus satanas Lenz (Boletaceae). Arch. Toxicol. Suppl. 1989, 13, 422–427. [Google Scholar] [PubMed]
- Zhao, K.; Wu, G.; Yang, Z.L. A new genus, Rubroboletus, to accommodate Boletus sinicus and its allies. Phytotaxa 2014, 188, 61–77. [Google Scholar] [CrossRef]
- Licastro, F.; Morini, M.C.; Kretz, O.; Dirheimer, G.; Creppy, E.E.; Stirpe, F. Mitogenic activity and immunological properties of bolesatine, a lectin isolated from the mushroom Boletus satanas Lenz. Int. J. Biochem. 1993, 25, 789–792. [Google Scholar] [CrossRef]
- Ng, T.B.; Wang, H.X. Flammin and velin: New ribosome inactivating polypeptides from the mushroom Flammulina velutipes. Peptides 2004, 25, 929–933. [Google Scholar] [CrossRef]
- Wang, H.X.; Ng, T.B. Flammulin: A novel ribosome-inactivating protein from fruiting bodies of the winter mushroom Flammulina velutipes. Biochem. Cell Biol. 2000, 78, 699–702. [Google Scholar] [CrossRef]
- Lam, S.K.; Ng, T.B. Hypsin, a novel thermostable ribosome-inactivating protein with antifungal and antiproliferative activities from fruiting bodies of the edible mushroom Hypsizigus marmoreus. Biochem. Biophys. Res. Commun. 2001, 285, 1071–1075. [Google Scholar] [CrossRef]
- Wang, H.X.; Ng, T.B. Isolation of pleuturegin, a novel ribosome-inactivating protein from fresh sclerotia of the edible mushroom Pleurotus tuber-regium. Biochem. Biophys. Res. Commun. 2001, 288, 718–721. [Google Scholar] [CrossRef]
- Lin, A.; Chen, C.K.; Chen, Y.J. Molecular action of tricholin, a ribosome-inactivating protein isolated from Trichoderma viride. Mol. Microbiol. 1991, 5, 3007–3013. [Google Scholar] [CrossRef]
- Wang, H.; Ng, T.B. Isolation and characterization of velutin, a novel low-molecular-weight ribosome-inactivating protein from winter mushroom (Flammulina velutipes) fruiting bodies. Life Sci. 2001, 68, 2151–2158. [Google Scholar] [CrossRef]
- Ng, T.B.; Lam, J.S.; Wong, J.H.; Lam, S.K.; Ngai, P.H.; Wang, H.X.; Chu, K.T.; Chan, W.Y. Differential abilities of the mushroom ribosome-inactivating proteins hypsin and velutin to perturb normal development of cultured mouse embryos. Toxicol. Vitr. 2010, 24, 1250–1257. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.M.; Xing, Y.M.; Guo, S.X. Molecular cloning and prokaryotic expression of a type II ribosome inactivating protein from Polyporus umbellatus. Yao Xue Xue Bao 2017, 52, 327–332. [Google Scholar] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]






| Enzyme | Source | Mr (Da) | N Terminal Sequence | Taxonomy NCBI: Txid | Ref. |
|---|---|---|---|---|---|
| Calcaelin | Calvatia caelata | 39,000 | 1 ANPIYNIDAF RV 12 | 1916349 | [34] |
| Lyophyllin | Lyophyllum shimeji | 20,000 | 1 ITFQGCSPAR QTVITNAITR ARADVRAAVS 30 * | 47721 | [35,36] |
| Marmorin | Hypsizygus marmoreus | 9567 | 1 AEGTLLGSRA TCESGNSMY 19 | 39966 | [37] |
| Mucoricin | Rhizopus delemar | 17,000 | 1 YFEEGRLFFI KSQFNGRVLD VEDGSTEDDA 30 * | 936053 | [38] |
| Volvarin | Volvariella valvacea | 29,000 | n.d. | 36659 | [39] |
| Enzyme | Source | Protein Yield (mg/kg) | IC50 Value (nM) | Ref. |
|---|---|---|---|---|
| Calcaelin | Calvatia caelata | 10.0 | 4.0 | [34] |
| Lyophyllin | Lyophyllum shimeji | 1.66 | 1.0 | [35,36] |
| Marmorin | Hypsizygus marmoreus | 2.2 | 0.7 | [37] |
| Mucoricin | Rhizopus delemar | n.r. | 17 | [38] |
| Volvarin | Volvariella valvacea | n.r. | 0.5 | [39] |
| Enzyme | Source | Mr (Da) | N Terminal Sequence | Taxonomy NCBI: Txid | Ref. |
|---|---|---|---|---|---|
| Bolesatine | Boletus satanas | 63,000 | 1 TWRIYLNNQT VKLALLLPNG 20 | 5370 | [32] |
| Flammin | Flammulina velutipes | 30,000 | 1 SPVIPANTFV AFRLYEVGIV PA 22 | 38945 | [46] |
| Flammulin | Flammulina velutipes | 40,000 | 1 APSHFSHPGV LADRAQIDFI XGKVNEGAEP WXSAYN 36 | 38945 | [47] |
| Hypsin | Flammulina velutipes | 20,000 | 1 ITFQGDLDAR QQVITNADTR RKRDVRAAVR 30 | 38945 | [48] |
| Pleuturegin | Pleurotus tuber-regium | 38,000 | 1 ARTQPGNIAP VGDFTLYPNA PRQGHIVA 28 | 716892 | [49] |
| Tricholin | Trichoderma viride | 14,200 | n.d. | 5547 | [50] |
| Velin | Flammulina velutipes | 19,000 | 1 SGSPLTQAQA EALLKPQGLA YSSGGNT 27 | 38945 | [46] |
| Velutin | Flammulina velutipes | 13,800 | 1 XHPDLFXXRP DNTASPKFED PRLNP 25 | 38945 | [51] |
| Enzyme | Source | Protein Yield (mg/kg) | IC50 Value (nM) | Ref. |
|---|---|---|---|---|
| Bolesatine | Boletus satanas | 0.23 * | 33 | [32] |
| Flammin | Flammulina velutipes | 1.28 | 1.4 | [46] |
| Flammulin | Flammulina velutipes | 0.44 | 0.25 | [47] |
| Hypsin | Flammulina velutipes | 10.0 | 7.0 | [48] |
| Pleuturegin | Pleurotus tuber-regium | n.r. | 0.5 | [49] |
| Tricholin | Trichoderma viride | n.r. | 630 # | [50] |
| Velin | Flammulina velutipes | 1.1 | 2.5 | [46] |
| Velutin | Flammulina velutipes | 16.28 | 0.29 | [51] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landi, N.; Hussain, H.Z.F.; Pedone, P.V.; Ragucci, S.; Di Maro, A. Ribotoxic Proteins, Known as Inhibitors of Protein Synthesis, from Mushrooms and Other Fungi According to Endo’s Fragment Detection. Toxins 2022, 14, 403. https://doi.org/10.3390/toxins14060403
Landi N, Hussain HZF, Pedone PV, Ragucci S, Di Maro A. Ribotoxic Proteins, Known as Inhibitors of Protein Synthesis, from Mushrooms and Other Fungi According to Endo’s Fragment Detection. Toxins. 2022; 14(6):403. https://doi.org/10.3390/toxins14060403
Chicago/Turabian StyleLandi, Nicola, Hafiza Z. F. Hussain, Paolo V. Pedone, Sara Ragucci, and Antimo Di Maro. 2022. "Ribotoxic Proteins, Known as Inhibitors of Protein Synthesis, from Mushrooms and Other Fungi According to Endo’s Fragment Detection" Toxins 14, no. 6: 403. https://doi.org/10.3390/toxins14060403