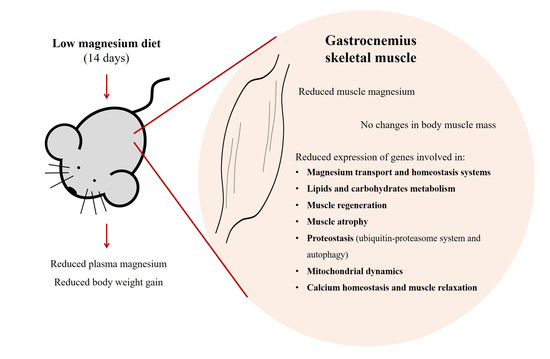
Magnesium Deficiency Alters Expression of Genes Critical for Muscle Magnesium Homeostasis and Physiology in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Mg2+ Analysis
2.3. Fiber Morphometry and Capillary Network
2.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
2.5. TaqMan Low-Density Array (TLDA)
2.6. Western Blot
2.7. Statistical Analysis
3. Results
3.1. Mg2+-Deficient Diet Reduces Muscle Mg2+ Concentrations but Does Not Affect Fiber Characteristics
3.2. Mg2+-Deficient Diet Alters Expression of Muscle MgTHs
3.3. Mild Mg2+ Deficiency Alters the Expression of Genes Important for Muscle Energy Metabolism and Regeneration
3.4. Mild Mg2+ Deficiency Alters Expression of Genes Important for Muscle Proteostasis
3.5. Mild Mg2+ Deficiency, Mitochondria, and Ca2+ Homeostasis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cazzola, R.; Della Porta, M.; Manoni, M.; Iotti, S.; Pinotti, L.; Maier, J.A. Going to the roots of reduced magnesium dietary intake: A tradeoff between climate changes and sources. Heliyon 2020, 6, e05390. [Google Scholar] [CrossRef] [PubMed]
- Lo Piano, F.; Corsonello, A.; Corica, F. Magnesium and elderly patient: The explored paths and the ones to be explored: A review. Magnes. Res. 2019, 32, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kisters, K.; Gröber, U. Magnesium and thiazide diuretics. Magnes. Res. 2018, 31, 143–145. [Google Scholar] [CrossRef]
- Hansen, B.A.; Bruserud, Ø. Hypomagnesemia in critically ill patients. J. Intensive Care 2018, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Arnaud, M.J. Update on the assessment of magnesium status. Br. J. Nutr. 2008, 99, S24–S36. [Google Scholar] [CrossRef] [Green Version]
- Witkowski, M.; Hubert, J.; Mazur, A. Methods of assessment of magnesium status in humans: A systematic review. Magnes. Res. 2011, 24, 163–180. [Google Scholar] [CrossRef] [Green Version]
- de Baaij, J.H.F.; Hoenderop, J.G.J.; Bindels, R.J.M. Magnesium in Man: Implications for Health and Disease. Physiol. Rev. 2015, 95, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Giménez-Mascarell, P.; González-Recio, I.; Fernández-Rodríguez, C.; Oyenarte, I.; Müller, D.; Martínez-Chantar, M.L.; Martínez-Cruz, L.A. Current Structural Knowledge on the CNNM Family of Magnesium Transport Mediators. Int. J. Mol. Sci. 2019, 20, 1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolisek, M.; Sponder, G.; Pilchova, I.; Cibulka, M.; Tatarkova, Z.; Werner, T.; Racay, P. Magnesium Extravaganza: A Critical Compendium of Current Research into Cellular Mg(2+) Transporters Other than TRPM6/7. Rev. Physiol. Biochem. Pharmacol. 2019, 176, 65–105. [Google Scholar] [CrossRef] [PubMed]
- Carvil, P.; Cronin, J. Magnesium and implications on muscle function. Strength Cond. J. 2010, 32, 48–54. [Google Scholar] [CrossRef]
- Coudy-Gandilhon, C.; Gueugneau, M.; Taillandier, D.; Combaret, L.; Polge, C.; Roche, F.; Barthélémy, J.-C.; Féasson, L.; Maier, J.A.; Mazur, A.; et al. Magnesium transport and homeostasis-related gene expression in skeletal muscle of young and old adults: Analysis of the transcriptomic data from the PROOF cohort Study. Magnes. Res. 2019, 32, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Gueugneau, M.; Coudy-Gandilhon, C.; Théron, L.; Meunier, B.; Barboiron, C.; Combaret, L.; Taillandier, D.; Polge, C.; Attaix, D.; Picard, B.; et al. Skeletal muscle lipid content and oxidative activity in relation to muscle fiber type in aging and metabolic syndrome. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 566–576. [Google Scholar] [CrossRef]
- Gueugneau, M.; Coudy-Gandilhon, C.; Meunier, B.; Combaret, L.; Taillandier, D.; Polge, C.; Attaix, D.; Roche, F.; Féasson, L.; Barthélémy, J.-C.; et al. Lower skeletal muscle capillarization in hypertensive elderly men. Exp. Gerontol. 2016, 76, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rondón, L.J.; Groenestege, W.M.T.; Rayssiguier, Y.; Mazur, A. Relationship between low magnesium status and TRPM6 expression in the kidney and large intestine. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R2001–R2007. [Google Scholar] [CrossRef] [PubMed]
- Ryazanova, L.V.; Rondon, L.J.; Zierler, S.; Hu, Z.; Galli, J.; Yamaguchi, T.P.; Mazur, A.; Fleig, A.; Ryazanov, A.G. TRPM7 is essential for Mg(2+) homeostasis in mammals. Nat. Commun. 2010, 1, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.Y.; Chaigne-Delalande, B.; Kanellopoulou, C.; Davis, J.C.; Matthews, H.F.; Douek, D.C.; Cohen, J.I.; Uzel, G.; Su, H.C.; Lenardo, M.J. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature 2011, 475, 471–476. [Google Scholar] [CrossRef]
- Blommaert, E.; Péanne, R.; Cherepanova, N.A.; Rymen, D.; Staels, F.; Jaeken, J.; Race, V.; Keldermans, L.; Souche, E.; Corveleyn, A.; et al. Mutations in MAGT1 lead to a glycosylation disorder with a variable phenotype. Proc. Natl. Acad. Sci. USA 2019, 116, 9865–9870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, C.; Masumiya, H.; Weisleder, N.; Matsuda, N.; Nishi, M.; Hwang, M.; Ko, J.-K.; Lin, P.; Thornton, A.; Zhao, X.; et al. MG53 nucleates assembly of cell membrane repair machinery. Nat. Cell Biol. 2009, 11, 56–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebert, S.M.; Bullard, S.A.; Basisty, N.; Marcotte, G.R.; Skopec, Z.P.; Dierdorff, J.M.; Al-Zougbi, A.; Tomcheck, K.C.; DeLau, A.D.; Rathmacher, J.A.; et al. Activating transcription factor 4 (ATF4) promotes skeletal muscle atrophy by forming a heterodimer with the transcriptional regulator C/EBPβ. J. Biol. Chem. 2020, 295, 2787–2803. [Google Scholar] [CrossRef] [Green Version]
- AlSudais, H.; Lala-Tabbert, N.; Wiper-Bergeron, N. CCAAT/Enhancer Binding Protein β inhibits myogenic differentiation via ID3. Sci. Rep. 2018, 8, 16613. [Google Scholar] [CrossRef]
- Ebert, S.M.; Al-Zougbi, A.; Bodine, S.C.; Adams, C.M. Skeletal Muscle Atrophy: Discovery of Mechanisms and Potential Therapies. Physiology 2019, 34, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Degens, H.; Li, M.; Salviati, L.; Lee, Y., II; Thompson, W.; Kirkland, J.L.; Sandri, M. Sarcopenia: Aging-Related Loss of Muscle Mass and Function. Physiol. Rev. 2019, 99, 427–511. [Google Scholar] [CrossRef] [PubMed]
- Gouspillou, G.; Hepple, R.T. Editorial: Mitochondria in Skeletal Muscle Health, Aging and Diseases. Front. Physiol. 2016, 7, 446. [Google Scholar] [CrossRef] [Green Version]
- Leberer, E.; Timms, B.G.; Campbell, K.P.; MacLennan, D.H. Purification, calcium binding properties, and ultrastructural localization of the 53,000- and 160,000 (sarcalumenin)-dalton glycoproteins of the sarcoplasmic reticulum. J. Biol. Chem. 1990, 265, 10118–10124. [Google Scholar] [CrossRef]
- Gueugneau, M.; Coudy-Gandilhon, C.; Gourbeyre, O.; Chambon, C.; Combaret, L.; Polge, C.; Taillandier, D.; Attaix, D.; Friguet, B.; Maier, A.B.; et al. Proteomics of muscle chronological ageing in post-menopausal women. BMC Genom. 2014, 15, 1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petermann-Rocha, F.; Chen, M.; Gray, S.R.; Ho, F.K.; Pell, J.P.; Celis-Morales, C. Factors associated with sarcopenia: A cross-sectional analysis using UK Biobank. Maturitas 2020, 133, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Ter Borg, S.; de Groot, L.C.P.G.M.; Mijnarends, D.M.; de Vries, J.H.M.; Verlaan, S.; Meijboom, S.; Luiking, Y.C.; Schols, J.M.G.A. Differences in Nutrient Intake and Biochemical Nutrient Status Between Sarcopenic and Nonsarcopenic Older Adults-Results From the Maastricht Sarcopenia Study. J. Am. Med. Dir. Assoc. 2016, 17, 393–401. [Google Scholar] [CrossRef] [Green Version]
- Orsso, C.E.; Tibaes, J.R.B.; Oliveira, C.L.P.; Rubin, D.A.; Field, C.J.; Heymsfield, S.B.; Prado, C.M.; Haqq, A.M. Low muscle mass and strength in pediatrics patients: Why should we care? Clin. Nutr. 2019, 38, 2002–2015. [Google Scholar] [CrossRef]
- Chubanov, V.; Ferioli, S.; Wisnowsky, A.; Simmons, D.G.; Leitzinger, C.; Einer, C.; Jonas, W.; Shymkiv, Y.; Bartsch, H.; Braun, A.; et al. Epithelial magnesium transport by TRPM6 is essential for prenatal development and adult survival. Elife 2016, 5. [Google Scholar] [CrossRef]
- Franken, G.A.C.; Seker, M.; Bos, C.; Siemons, L.A.H.; van der Eerden, B.C.J.; Christ, A.; Hoenderop, J.G.J.; Bindels, R.J.M.; Müller, D.; Breiderhoff, T.; et al. Cyclin M2 (CNNM2) knockout mice show mild hypomagnesaemia and developmental defects. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Milan, G.; Romanello, V.; Pescatore, F.; Armani, A.; Paik, J.H.; Frasson, L.; Seydel, A.; Zhao, J.; Abraham, R.; Goldberg, A.L.; et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat. Commun. 2015, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Morakinyo, A.O.; Samuel, T.A.; Adekunbi, D.A. Magnesium upregulates insulin receptor and glucose transporter-4 in streptozotocin-nicotinamide-induced type-2 diabetic rats. Endocr. Regul. 2018, 52, 6–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sewter, C.; Berger, D.; Considine, R.V.; Medina, G.; Rochford, J.; Ciaraldi, T.; Henry, R.; Dohm, L.; Flier, J.S.; O’Rahilly, S.; et al. Human obesity and type 2 diabetes are associated with alterations in SREBP1 isoform expression that are reproduced ex vivo by tumor necrosis factor-alpha. Diabetes 2002, 51, 1035–1041. [Google Scholar] [CrossRef] [Green Version]
- Bosma, M.; Hesselink, M.K.C.; Sparks, L.M.; Timmers, S.; Ferraz, M.J.; Mattijssen, F.; van Beurden, D.; Schaart, G.; de Baets, M.H.; Verheyen, F.K.; et al. Perilipin 2 improves insulin sensitivity in skeletal muscle despite elevated intramuscular lipid levels. Diabetes 2012, 61, 2679–2690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brault, J.J.; Abraham, K.A.; Terjung, R.L. Muscle creatine uptake and creatine transporter expression in response to creatine supplementation and depletion. J. Appl. Physiol. 2003, 94, 2173–2180. [Google Scholar] [CrossRef] [Green Version]
- Stockebrand, M.; Sasani, A.; Das, D.; Hornig, S.; Hermans-Borgmeyer, I.; Lake, H.A.; Isbrandt, D.; Lygate, C.A.; Heerschap, A.; Neu, A.; et al. A Mouse Model of Creatine Transporter Deficiency Reveals Impaired Motor Function and Muscle Energy Metabolism. Front. Physiol. 2018, 9, 773. [Google Scholar] [CrossRef]
- Filadi, R.; Pendin, D.; Pizzo, P. Mitofusin 2: From functions to disease. Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, S.; Romeo, V.; Locatelli, L.; Cazzaniga, A.; Maier, J.A.M. TRPM7 and MagT1 in the osteogenic differentiation of human mesenchymal stem cells in vitro. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Yue, J.; Jin, S.; Gu, S.; Sun, R.; Liang, Q. High concentration magnesium inhibits extracellular matrix calcification and protects articular cartilage via Erk/autophagy pathway. J. Cell. Physiol. 2019, 234, 23190–23201. [Google Scholar] [CrossRef]







| Primers | Forward | Reverse |
|---|---|---|
| TRPM6 | GACAGTCTAAGCACCTTTTC | AAGCTTGTACCCTTCAGTAG |
| MagT1 | GATGGGCTTTTGCAGCTTTGT | GCAATACACATCATCCTTCGCT |
| MRS2 | GGTGATGTGCTCCGGTTTAGA | TGGCCTGGAGTGCTAACTCAT |
| Slc41a1 | TACTGGCCCTACTCCTTCTCC | GGGACTCAATCACTACCACCTC |
| Slc41a2 | CTTCAGCAAGAGATCAGAGCC | CCAGCATAGTCATCGTACTTGG |
| Slc41a3 | CTCAGCCTTGAGTTCCGCTTT | GCAGGATAGGTATGGCGACC |
| CNNM1 | GTAGGGTCACAACCTACATC | CATGACATACACAGAAGAGG |
| CNNM2 | AAGTGGCCCACCGTGAAAG | CGCTTCTACTTCTGTTGCTAGG |
| CNNM3 | GACTCCGGCACTGTCCTAGA | AGTGGATGGTTGTAGAAGCGG |
| CNNM4 | CTGCACATCCTTCTCGTTATGG | TGCGAGCATACTTTCTCTCCTT |
| Acvr2a | Activin A Receptor Type 2a | Map1lc3b | Microtubule-Associated Protein 1 Light Chain 3 Beta |
| Acvr2b | Activin A Receptor Type 2b | Mdm2 | Mouse double minute 2 homolog |
| Asb2 | Ankyrin Repeat And SOCS Box Containing 2 | Mef2c | Myocyte Enhancer Factor 2C |
| Atf4 | Activating Transcription Factor 4 | Mfn1 | Mitofusin 1 |
| Atg10 | Autophagy Related 10 | Mfn2 | Mitofusin 2 |
| Atg12 | Autophagy Related 12 | Mief1 | Mitochondrial Elongation Factor 1 |
| Atg16l1 | Autophagy Related 16 Like 1 | Mstn | Myostatin |
| Atg3 | Autophagy Related 3 | Myf5 | Myogenic Factor 5 |
| Atg5 | Autophagy Related 5 | Myod | Myogenic Differentiation 1 |
| Atg7 | Autophagy Related 7 | Myog | Myogenin |
| Atp2a1 | ATPase Sarcoplasmic/Endoplasmic Reticulum Ca2+ Transporting 1 | Nbr1 | NBR1 Autophagy Cargo Receptor |
| Becn1 | Beclin 1 | Nrf1 | Nuclear Respiratory Factor 1 |
| Bnip3 | BCL2/adenovirus E1B Interacting Protein 3 | Opa1 | Mitochondrial dynamin like GTPase |
| Capn1 | Calpain 1 | Park2 | Parkin RBR E3 Ubiquitin Protein Ligase |
| Capn2 | Calpain 2 | Pik3c3 | Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3 |
| Capn3 | Calpain 3 | Pink1 | PTEN Induced Kinase 1 |
| Casq1 | Calsequestrin 1 | Plin2 | Perilipin 2 |
| Cdkn1a | Cyclin Dependent Kinase Inhibitor 1A | Plin5 | Perilipin 5 |
| Chac1 | Glutathione-specific gamma-glutamylcyclotransferase 1 | Ppargc1a | Peroxisome Proliferative Activated Receptor Gamma Coactivator 1 Alpha |
| CNNM2 | Cyclin and CBS Domain Divalent Metal Cation Transport Mediator 2 | Rhot1 | Ras Homolog Family Member T1 |
| CNNM4 | Cyclin and CBS Domain Divalent Metal Cation Transport Mediator 4 | Ryr1 | Ryanodine Receptor 1 |
| Cox1 | Mitochondrially Encoded Cytochrome C Oxidase I | Slc27a1 | Solute Carrier Family 27 Member 1 |
| Creb1 | CAMP Responsive Element Binding Protein 1 | Slc2a1 | Solute Carrier Family 2 Member 1 |
| Cs | citrate synthase | Slc2a4 | Solute Carrier Family 2 Member 4 |
| Ctsl | Cathepsin L | Slc6a8 | Solute Carrier Family 6 Member 8 |
| Ddit3 | DNA Damage Inducible Transcript 3 | Slc41a1 | Solute Carrier family 41 member 1 |
| Dnm1l | Dynamin 1 Like | Sqstm1 | Sequestosome 1 |
| Eif4ebp1 | Eukaryotic Translation Initiation Factor 4E Binding Protein 1 | Srebf1 | Sterol Regulatory Element Binding Transcription Factor 1 |
| Fabp3 | Fatty Acid Binding Protein 3 | Srebf2 | Sterol Regulatory Element Binding Transcription Factor 2 |
| Fbxo21 | F-Box Protein 21 | Srl | Sarcalumenin |
| Fbxo30 | F-Box Protein 30 | Tfam | Transcription Factor A, Mitochondrial |
| Fbxo31 | F-Box Protein 31 | Trim63 | Tripartite Motif-containing 63 |
| Fbxo32 | F-Box Protein 32 | Trim72 | Tripartite Motif-containing 72 |
| Fis1 | Fission, Mitochondrial 1 | TRPM6 | Transient Receptor Potential cation channel subfamily M member 6 |
| Foxo3 | Forkhead Box O3 | Ube2b | Ubiquitin Conjugating Enzyme E2 B |
| Fst | Follistatin | Ube2e1 | Ubiquitin-Conjugating Enzyme E2 E1 |
| Fundc1 | FUN14 Domain Containing 1 | Ube2g1 | Ubiquitin-Conjugating Enzyme E2G 1 |
| Gabarapl1v | Gamma-Aminobutyric Acid A Receptor-Associated Protein-Like 1 | Ube2j1 | Ubiquitin-Conjugating Enzyme E2J 1 |
| Gadd45a | Growth Arrest and DNA Damage Inducible Alpha | Ube2j2 | Ubiquitin-Conjugating Enzyme E2J 2 |
| Gapdh | Glyceraldehyde-3-Phosphate Dehydrogenase | Ube2l3 | Ubiquitin-Conjugating Enzyme E2L 3 |
| Lamp2 | Lysosomal-Associated Membrane Protein 2 | Ulk1 | Unc-51 Like Autophagy Activating Kinase 1 |
| MagT1 | Magnesium Transporter 1 | Zeb1 | Zinc Finger E-Box Binding Homeobox 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayle, D.; Coudy-Gandilhon, C.; Gueugneau, M.; Castiglioni, S.; Zocchi, M.; Maj-Zurawska, M.; Palinska-Saadi, A.; Mazur, A.; Béchet, D.; Maier, J.A. Magnesium Deficiency Alters Expression of Genes Critical for Muscle Magnesium Homeostasis and Physiology in Mice. Nutrients 2021, 13, 2169. https://doi.org/10.3390/nu13072169
Bayle D, Coudy-Gandilhon C, Gueugneau M, Castiglioni S, Zocchi M, Maj-Zurawska M, Palinska-Saadi A, Mazur A, Béchet D, Maier JA. Magnesium Deficiency Alters Expression of Genes Critical for Muscle Magnesium Homeostasis and Physiology in Mice. Nutrients. 2021; 13(7):2169. https://doi.org/10.3390/nu13072169
Chicago/Turabian StyleBayle, Dominique, Cécile Coudy-Gandilhon, Marine Gueugneau, Sara Castiglioni, Monica Zocchi, Magdalena Maj-Zurawska, Adriana Palinska-Saadi, André Mazur, Daniel Béchet, and Jeanette A. Maier. 2021. "Magnesium Deficiency Alters Expression of Genes Critical for Muscle Magnesium Homeostasis and Physiology in Mice" Nutrients 13, no. 7: 2169. https://doi.org/10.3390/nu13072169