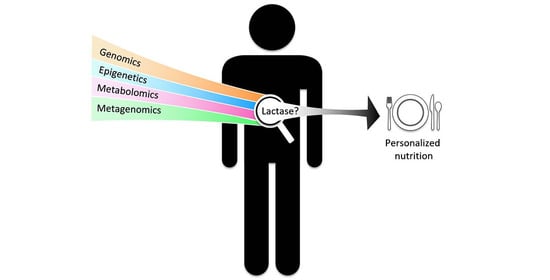
Development of Personalized Nutrition: Applications in Lactose Intolerance Diagnosis and Management
Abstract
:1. Introduction
2. Lactose Intolerance: Current Clinical Management
2.1. Physiology and Pathophysiology of Lactose Intolerance
2.2. Types of Lactose Intolerance
2.3. Diagnosis of Lactose Intolerance
2.4. Management of Lactose Intolerance
3. Prolonged Health Implications of Lactose Intolerance
3.1. Lactose Intolerance and Bone Health
3.2. Lactose Intolerance and Non-Communicable Chronic Diseases
3.3. Lactose Intolerance and Colonic Health
4. Application of “Omics” Tools to Research on Lactose Intolerance
4.1. Application of Genomics for Genetic Testing of Lactase Persistence
4.2. Epigenetics of Lactase Regulation over the Lifespan
4.3. Metagenomics, Lactose Intolerance, and the Gut Microbiota
4.4. Current and Future Perspectives of Nutritional Metabolomics
5. Lactose Intolerance as a Research Model towards Personalized Nutrition
5.1. Personalized Diets and Population Health
5.2. Personalized Nutrition for Health Optimization, and Disease Management
5.3. Food Market, Product Innovation, and Affordability
5.4. Challenges for Developing Personalized Nutrition
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaput, J.; Kussmann, M.; Radonjic, M.; Virgili, F.; Perozzi, G. Human nutrition, environment, and health. Genes Nutr. 2015, 10, 489. [Google Scholar] [CrossRef] [PubMed]
- Walther, B.; Lett, A.M.; Bordoni, A.; Tomás-Cobos, L.; Nieto, J.A.; Dupont, D.; Danesi, F.; Shahar, D.R.; Echaniz, A.; Re, R.; et al. GutSelf: Interindividual Variability in the Processing of Dietary Compounds by the Human Gastrointestinal Tract. Mol. Nutr. Food Res. 2019, 63, e1900677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised nutrition and health. BMJ 2018, 361, bmj.k2173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bush, C.L.; Blumberg, J.B.; El-Sohemy, A.; Minich, D.M.; Ordovás, J.M.; Reed, D.G.; Behm, V.A.Y. Toward the Definition of Personalized Nutrition: A Proposal by The American Nutrition Association. J. Am. Coll. Nutr. 2020, 39, 5–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattar, R.; de Campos Mazo, D.F.; Carrilho, F.J. Lactose intolerance: Diagnosis, genetic, and clinical factors. Clin. Exp. Gastroenterol. 2012, 5, 113–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiley, A.S. Lactose intolerance. Evol. Med. Public Health 2020, 2020, 47–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storhaug, C.L.; Fosse, S.K.; Fadnes, L.T. Country, regional, and global estimates for lactose malabsorption in adults: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2017, 2, 738–746. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.D.; Lawlor, D.A.; Timpson, N.J.; Baban, J.; Kiessling, M.; Day, I.N.M.; Ebrahim, S. Lactase persistence-related genetic variant: Population substructure and health outcomes. Eur. J. Hum. Genet. 2009, 17, 357–367. [Google Scholar] [CrossRef] [Green Version]
- Comerford, K.B.; Pasin, G. Gene-Dairy Food Interactions and Health Outcomes: A Review of Nutrigenetic Studies. Nutrients 2017, 9, 710. [Google Scholar] [CrossRef] [Green Version]
- Itan, Y.; Jones, B.L.; Ingram, C.J.; Swallow, D.M.; Thomas, M.G. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol. Biol. 2010, 10, 36. [Google Scholar] [CrossRef] [Green Version]
- Tishkoff, S.A.; Reed, F.A.; Ranciaro, A.; Voight, B.F.; Babbitt, C.C.; Silverman, J.S.; Powell, K.; Mortensen, H.M.; Hirbo, J.B.; Osman, M.; et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 2007, 39, 31–40. [Google Scholar] [CrossRef]
- Luca, F.; Perry, G.H.; Di Rienzo, A. Evolutionary adaptations to dietary changes. Annu. Rev. Nutr. 2010, 30, 291–314. [Google Scholar] [CrossRef] [Green Version]
- Gerbault, P.; Liebert, A.; Itan, Y.; Powell, A.; Currat, M.; Burger, J.; Swallow, D.M.; Thomas, M.G. Evolution of lactase persistence: An example of human niche construction. Philos. Trans. R. Soc. Lond B Biol. Sci. 2011, 366, 863–877. [Google Scholar] [CrossRef] [Green Version]
- Itan, Y.; Powell, A.; Beaumont, M.A.; Burger, J.; Thomas, M.G. The origins of lactase persistence in Europe. PLoS Comput. Biol. 2009, 5, e1000491. [Google Scholar] [CrossRef] [Green Version]
- Segurel, L.; Guarino-Vignon, P.; Marchi, N.; Lafosse, S.; Laurent, R.; Bon, C.; Fabre, A.; Hegay, T.; Heyer, E. Why and when was lactase persistence selected for? Insights from Central Asian herders and ancient DNA. PLoS Biol. 2020, 18, e3000742. [Google Scholar] [CrossRef]
- Ségurel, L.; Bon, C. On the Evolution of Lactase Persistence in Humans. Annu. Rev. Genom. Hum. Genet. 2017, 18, 297–319. [Google Scholar] [CrossRef]
- Lomer, M.C.; Parkes, G.C.; Sanderson, J.D. Review article: Lactose intolerance in clinical practice-myths and realities. Aliment. Pharmacol. 2008, 27, 93–103. [Google Scholar] [CrossRef]
- Ingram, C.J.E.; Mulcare, C.A.; Itan, Y.; Thomas, M.G.; Swallow, D.M. Lactose digestion and the evolutionary genetics of lactase persistence. Hum. Genet. 2009, 124, 579–591. [Google Scholar] [CrossRef]
- Kuchay, R.A.H. New insights into the molecular basis of lactase non-persistence/persistence: A brief review. Drug Discov. Ther. 2020, 14, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Forsgård, R.A. Lactose digestion in humans: Intestinal lactase appears to be constitutive whereas the colonic microbiome is adaptable. Am. J. Clin. Nutr. 2019, 110, 273–279. [Google Scholar] [CrossRef]
- Misselwitz, B.; Butter, M.; Verbeke, K.; Fox, M.R. Update on lactose malabsorption and intolerance: Pathogenesis, diagnosis and clinical management. Gut 2019, 68, 2080–2091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, T.; Venema, K.; Priebe, M.G.; Welling, G.W.; Brummer, R.J.; Vonk, R.J. The role of colonic metabolism in lactose intolerance. Eur. J. Clin. Investig. 2008, 38, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Misselwitz, B.; Dai, N.; Fox, M. Lactose Intolerance in Adults: Biological Mechanism and Dietary Management. Nutrients 2015, 7, 8020–8035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szilagyi, A. Adult lactose digestion status and effects on disease. Can. J. Gastroenterol. Hepatol. 2015, 29, 149–156. [Google Scholar] [CrossRef]
- Swallow, D.M. Genetics of lactase persistence and lactose intolerance. Annu. Rev. Genet. 2003, 37, 197–219. [Google Scholar] [CrossRef]
- Ingram, C.J.; Elamin, M.F.; Mulcare, C.A.; Weale, M.E.; Tarekegn, A.; Raga, T.O.; Bekele, E.; Elamin, F.M.; Thomas, M.G.; Bradman, N.; et al. A novel polymorphism associated with lactose tolerance in Africa: Multiple causes for lactase persistence? Hum. Genet. 2007, 120, 779–788. [Google Scholar] [CrossRef]
- Enattah, N.S.; Sahi, T.; Savilahti, E.; Terwilliger, J.D.; Peltonen, L.; Järvelä, I. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 2002, 30, 233–237. [Google Scholar] [CrossRef]
- Santonocito, C.; Scapaticci, M.; Guarino, D.; Annicchiarico, E.B.; Lisci, R.; Penitente, R.; Gasbarrini, A.; Zuppi, C.; Capoluongo, E. Lactose intolerance genetic testing: Is it useful as routine screening? Results on 1426 south-central Italy patients. Clin. Chim. Acta 2015, 439, 14–17. [Google Scholar] [CrossRef]
- Tomczonek-Moruś, J.; Wojtasik, A.; Zeman, K.; Smolarz, B.; Bąk-Romaniszyn, L. 13910C>T and 22018G>A LCT gene polymorphisms in diagnosing hypolactasia in children. United Eur. Gastroenterol. J. 2019, 7, 210–216. [Google Scholar] [CrossRef] [Green Version]
- Enattah, N.-S.; Kuokkanen, M.; Forsblom, C.; Natah, S.; Oksanen, A.; Jarvela, I.; Peltonen, L.; Savilahti, E. Correlation of intestinal disaccharidase activities with the C/T-13910 variant and age. World J. Gastroenterol. 2007, 13, 3508–3512. [Google Scholar] [CrossRef] [Green Version]
- Gugatschka, M.; Dobnig, H.; Fahrleitner-Pammer, A.; Pietschmann, P.; Kudlacek, S.; Strele, A.; Obermayer-Pietsch, B. Molecularly-defined lactose malabsorption, milk consumption and anthropometric differences in adult males. Qjm 2005, 98, 857–863. [Google Scholar] [CrossRef]
- Matthews, S.B.; Waud, J.P.; Roberts, A.G.; Campbell, A.K. Systemic lactose intolerance: A new perspective on an old problem. Postgrad Med. J. 2005, 81, 167–173. [Google Scholar] [CrossRef] [Green Version]
- Troelsen, J.T. Adult-type hypolactasia and regulation of lactase expression. Biochim. Biophys. Acta 2005, 1723, 19–32. [Google Scholar] [CrossRef]
- Szilagyi, A.; Ishayek, N. Lactose Intolerance, Dairy Avoidance, and Treatment Options. Nutrients 2018, 10, 1994. [Google Scholar] [CrossRef] [Green Version]
- Walsh, J.; Meyer, R.; Shah, N.; Quekett, J.; Fox, A.T. Differentiating milk allergy (IgE and non-IgE mediated) from lactose intolerance: Understanding the underlying mechanisms and presentations. Br. J. Gen. Pr. 2016, 66, e609–e611. [Google Scholar] [CrossRef] [Green Version]
- Mishkin, B.; Yalovsky, M.; Mishkin, S. Increased prevalence of lactose malabsorption in Crohn’s disease patients at low risk for lactose malabsorption based on ethnic origin. Am. J. Gastroenterol. 1997, 92, 1148–1153. [Google Scholar]
- Szilagyi, A.; Galiatsatos, P.; Xue, X. Systematic review and meta-analysis of lactose digestion, its impact on intolerance and nutritional effects of dairy food restriction in inflammatory bowel diseases. Nutr. J. 2016, 15, 67. [Google Scholar] [CrossRef] [Green Version]
- Hasan, N.; Zainaldeen, M.; Almadhoob, F.; Yusuf, M.; Fredericks, S. Knowledge of lactose intolerance among clinicians. Gastroenterol. Insights 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Martínez Vázquez, S.E.; Nogueira de Rojas, J.R.; Remes Troche, J.M.; Coss Adame, E.; Rivas Ruíz, R.; Uscanga Domínguez, L.F. The importance of lactose intolerance in individuals with gastrointestinal symptoms. Rev. Gastroenterol. México 2020, 85, 321–331. [Google Scholar] [CrossRef]
- Lomer, M.C. Review article: The aetiology, diagnosis, mechanisms and clinical evidence for food intolerance. Aliment. Pharmacol. Ther. 2015, 41, 262–275. [Google Scholar] [CrossRef]
- Lule, V.K.; Garg, S.; Tomar, S.K.; Khedkar, C.D.; Nalage, D.N. Food Intolerance: Lactose Intolerance. In Encyclopedia of Food and Health; Caballero, B., Finglas, P.M., Toldrá, F., Eds.; Academic Press: Oxford, UK, 2016; pp. 43–48. [Google Scholar] [CrossRef]
- Di Rienzo, T.; D’Angelo, G.; D’Aversa, F.; Campanale, M.C.; Cesario, V.; Montalto, M.; Gasbarrini, A.; Ojetti, V. Lactose intolerance: From diagnosis to correct management. Eur. Rev. Med. Pharmacol. Sci. 2013, 17 (Suppl. 2), 18–25. [Google Scholar] [PubMed]
- Robles, L.; Priefer, R. Lactose Intolerance: What Your Breath Can Tell You. Diagnostics 2020, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Law, D.; Conklin, J.; Pimentel, M. Lactose intolerance and the role of the lactose breath test. Am. J. Gastroenterol. 2010, 105, 1726–1728. [Google Scholar] [CrossRef] [PubMed]
- Usai-Satta, P.; Scarpa, M.; Oppia, F.; Cabras, F. Lactose malabsorption and intolerance: What should be the best clinical management? World J. Gastrointest. Pharmacol. Ther. 2012, 3, 29–33. [Google Scholar] [CrossRef]
- Furnari, M.; Bonfanti, D.; Parodi, A.; Franzè, J.; Savarino, E.; Bruzzone, L.; Moscatelli, A.; Di Mario, F.; Dulbecco, P.; Savarino, V. A comparison between lactose breath test and quick test on duodenal biopsies for diagnosing lactase deficiency in patients with self-reported lactose intolerance. J. Clin. Gastroenterol. 2013, 47, 148–152. [Google Scholar] [CrossRef]
- Hovde, Ø.; Farup, P.G. A comparison of diagnostic tests for lactose malabsorption—Which one is the best? BMC Gastroenterol. 2009, 9, 82. [Google Scholar] [CrossRef] [Green Version]
- Aragón, J.J.; Hermida, C.; Martínez-Costa, O.H.; Sánchez, V.; Martín, I.; Sánchez, J.J.; Codoceo, R.; Cano, J.M.; Cano, A.; Crespo, L.; et al. Noninvasive diagnosis of hypolactasia with 4-Galactosylxylose (Gaxilose): A multicentre, open-label, phase IIB-III nonrandomized trial. J. Clin. Gastroenterol. 2014, 48, 29–36. [Google Scholar] [CrossRef]
- Domínguez Jiménez, J.L.; Fernández Suárez, A.; Muñoz Colmenero, A.; Fatela Cantillo, D.; López Pelayo, I. Primary hypolactasia diagnosis: Comparison between the gaxilose test, shortened lactose tolerance test, and clinical parameters corresponding to the C/T-13910 polymorphism. Clin. Nutr. 2017, 36, 471–476. [Google Scholar] [CrossRef]
- Bayless, T.M.; Brown, E.; Paige, D.M. Lactase Non-persistence and Lactose Intolerance. Curr. Gastroenterol. Rep. 2017, 19, 23. [Google Scholar] [CrossRef]
- Gille, D.; Walther, B.; Badertscher, R.; Bosshart, A.; Brügger, C.; Brühlhart, M.; Gauch, R.; Noth, P.; Vergères, G.; Egger, L. Detection of lactose in products with low lactose content. Int. Dairy J. 2018, 83, 17–19. [Google Scholar] [CrossRef]
- Dekker, P.J.T.; Koenders, D.; Bruins, M.J. Lactose-Free Dairy Products: Market Developments, Production, Nutrition and Health Benefits. Nutrients 2019, 11, 551. [Google Scholar] [CrossRef] [Green Version]
- Facioni, M.S.; Raspini, B.; Pivari, F.; Dogliotti, E.; Cena, H. Nutritional management of lactose intolerance: The importance of diet and food labelling. J. Transl. Med. 2020, 18, 260. [Google Scholar] [CrossRef]
- McSweeney, P.L.H. Biochemistry of cheese ripening. Int. J. Dairy Technol. 2004, 57, 127–144. [Google Scholar] [CrossRef]
- Savaiano, D.A. Lactose digestion from yogurt: Mechanism and relevance. Am. J. Clin. Nutr. 2014, 99, 1251s–1255s. [Google Scholar] [CrossRef] [Green Version]
- Van der Merwe, J.; Steenekamp, J.; Steyn, D.; Hamman, J. The Role of Functional Excipients in Solid Oral Dosage Forms to Overcome Poor Drug Dissolution and Bioavailability. Pharmaceutics 2020, 12, 393. [Google Scholar] [CrossRef]
- Mill, D.; Dawson, J.; Johnson, J.L. Managing acute pain in patients who report lactose intolerance: The safety of an old excipient re-examined. Adv. Drug Saf. 2018, 9, 227–235. [Google Scholar] [CrossRef] [Green Version]
- Sethi, S.; Tyagi, S.K.; Anurag, R.K. Plant-based milk alternatives an emerging segment of functional beverages: A review. J. Food Sci. Technol. 2016, 53, 3408–3423. [Google Scholar] [CrossRef]
- Harju, M.; Kallioinen, H.; Tossavainen, O. Lactose hydrolysis and other conversions in dairy products: Technological aspects. Int. Dairy J. Int. Dairy J. 2012, 22. [Google Scholar] [CrossRef]
- Vyas, H.K.; Tong, P.S. Process for Calcium Retention During Skim Milk Ultrafiltration. J. Dairy Sci. 2003, 86, 2761–2766. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, M.; Eaton, T.K.; Sermet, O.M.; Savaiano, D.A. Milk Containing A2 β-Casein ONLY, as a Single Meal, Causes Fewer Symptoms of Lactose Intolerance than Milk Containing A1 and A2 β-Caseins in Subjects with Lactose Maldigestion and Intolerance: A Randomized, Double-Blind, Crossover Trial. Nutrients 2020, 12, 3855. [Google Scholar] [CrossRef]
- Jianqin, S.; Leiming, X.; Lu, X.; Yelland, G.W.; Ni, J.; Clarke, A.J. Effects of milk containing only A2 beta casein versus milk containing both A1 and A2 beta casein proteins on gastrointestinal physiology, symptoms of discomfort, and cognitive behavior of people with self-reported intolerance to traditional cows’ milk. Nutr. J. 2016, 15, 35. [Google Scholar] [CrossRef] [Green Version]
- Suri, S.; Kumar, V.; Prasad, R.; Tanwar, B.; Goyal, A.; Kaur, S.; Gat, Y.; Kumar, A.; Kaur, J.; Singh, D. Considerations for development of lactose-free food. J. Nutr. Intermed. Metab. 2019, 15, 27–34. [Google Scholar] [CrossRef]
- Montalto, M.; Nucera, G.; Santoro, L.; Curigliano, V.; Vastola, M.; Covino, M.; Cuoco, L.; Manna, R.; Gasbarrini, A.; Gasbarrini, G. Effect of exogenous beta-galactosidase in patients with lactose malabsorption and intolerance: A crossover double-blind placebo-controlled study. Eur. J. Clin. Nutr. 2005, 59, 489–493. [Google Scholar] [CrossRef] [Green Version]
- Montalto, M.; Curigliano, V.; Santoro, L.; Vastola, M.; Cammarota, G.; Manna, R.; Gasbarrini, A.; Gasbarrini, G. Management and treatment of lactose malabsorption. World J. Gastroenterol. 2006, 12, 187–191. [Google Scholar] [CrossRef]
- Zheng, X.; Chu, H.; Cong, Y.; Deng, Y.; Long, Y.; Zhu, Y.; Pohl, D.; Fried, M.; Dai, N.; Fox, M. Self-reported lactose intolerance in clinic patients with functional gastrointestinal symptoms: Prevalence, risk factors, and impact on food choices. Neurogastroenterol. Motil. 2015, 27, 1138–1146. [Google Scholar] [CrossRef]
- Laing, B.B.; Lim, A.G.; Ferguson, L.R. A Personalised Dietary Approach-A Way Forward to Manage Nutrient Deficiency, Effects of the Western Diet, and Food Intolerances in Inflammatory Bowel Disease. Nutrients 2019, 11, 1532. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, E.E.; Fomon, S.J. Lactose enhances mineral absorption in infancy. J. Pediatr. Gastroenterol. Nutr. 1983, 2, 288–294. [Google Scholar] [CrossRef]
- Abrams, S.A.; Griffin, I.J.; Davila, P.M. Calcium and zinc absorption from lactose-containing and lactose-free infant formulas. Am. J. Clin. Nutr. 2002, 76, 442–446. [Google Scholar] [CrossRef] [Green Version]
- Romero-Velarde, E.; Delgado-Franco, D.; García-Gutiérrez, M.; Gurrola-Díaz, C.; Larrosa-Haro, A.; Montijo-Barrios, E.; Muskiet, F.A.J.; Vargas-Guerrero, B.; Geurts, J. The Importance of Lactose in the Human Diet: Outcomes of a Mexican Consensus Meeting. Nutrients 2019, 11, 2737. [Google Scholar] [CrossRef] [Green Version]
- Augustin, L.S.; Kendall, C.W.; Jenkins, D.J.; Willett, W.C.; Astrup, A.; Barclay, A.W.; Björck, I.; Brand-Miller, J.C.; Brighenti, F.; Buyken, A.E.; et al. Glycemic index, glycemic load and glycemic response: An International Scientific Consensus Summit from the International Carbohydrate Quality Consortium (ICQC). Nutr. Metab. Cardiovasc Dis. 2015, 25, 795–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rytz, A.; Adeline, D.; Lê, K.A.; Tan, D.; Lamothe, L.; Roger, O.; Macé, K. Predicting Glycemic Index and Glycemic Load from Macronutrients to Accelerate Development of Foods and Beverages with Lower Glucose Responses. Nutrients 2019, 11, 1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nickerson, K.P.; Chanin, R.; McDonald, C. Deregulation of intestinal anti-microbial defense by the dietary additive, maltodextrin. Gut Microbes 2015, 6, 78–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clouard, C.; Le Bourgot, C.; Respondek, F.; Bolhuis, J.E.; Gerrits, W.J.J. A milk formula containing maltodextrin, vs. lactose, as main carbohydrate source, improves cognitive performance of piglets in a spatial task. Sci. Rep. 2018, 8, 9433. [Google Scholar] [CrossRef]
- Clouard, C.; Lannuzel, C.; Bourgot, C.L.; Gerrits, W.J.J. Lactose and Digestible Maltodextrin in Milk Replacers Differently Affect Energy Metabolism and Substrate Oxidation: A Calorimetric Study in Piglets. J. Nutr. 2020, 150, 3114–3122. [Google Scholar] [CrossRef]
- Van der Reijden, O.L.; Zimmermann, M.B.; Galetti, V. Iodine in dairy milk: Sources, concentrations and importance to human health. Best Pr. Res. Clin. Endocrinol. Metab. 2017, 31, 385–395. [Google Scholar] [CrossRef]
- Nohr, D.; Biesalski, H. Vitamins in Milk and Dairy Products: B-Group Vitamins. Adv. Dairy Chem. 2009, 3. [Google Scholar] [CrossRef]
- Melse-Boonstra, A. Bioavailability of Micronutrients From Nutrient-Dense Whole Foods: Zooming in on Dairy, Vegetables, and Fruits. Front. Nutr. 2020, 7. [Google Scholar] [CrossRef]
- Laaksonen, M.M.; Impivaara, O.; Sievänen, H.; Viikari, J.S.; Lehtimäki, T.J.; Lamberg-Allardt, C.J.; Kärkkäinen, M.U.; Välimäki, M.; Heikkinen, J.; Kröger, L.M.; et al. Associations of genetic lactase non-persistence and sex with bone loss in young adulthood. Bone 2009, 44, 1003–1009. [Google Scholar] [CrossRef]
- Suchy, F.J.; Brannon, P.M.; Carpenter, T.O.; Fernandez, J.R.; Gilsanz, V.; Gould, J.B.; Hall, K.; Hui, S.L.; Lupton, J.; Mennella, J.; et al. NIH consensus development conference statement: Lactose intolerance and health. NIH Consens. State Sci. Statements 2010, 27, 1–27. [Google Scholar]
- Obermayer-Pietsch, B.M.; Bonelli, C.M.; Walter, D.E.; Kuhn, R.J.; Fahrleitner-Pammer, A.; Berghold, A.; Goessler, W.; Stepan, V.; Dobnig, H.; Leb, G.; et al. Genetic predisposition for adult lactose intolerance and relation to diet, bone density, and bone fractures. J. Bone Miner. Res. 2004, 19, 42–47. [Google Scholar] [CrossRef]
- Corazza, G.R.; Benati, G.; Di Sario, A.; Tarozzi, C.; Strocchi, A.; Passeri, M.; Gasbarrini, G. Lactose intolerance and bone mass in postmenopausal Italian women. Br. J. Nutr. 1995, 73, 479–487. [Google Scholar] [CrossRef]
- Hallkvist, O.M.; Johansson, J.; Nordström, A.; Nordström, P.; Hult, A. Dairy product intake and bone properties in 70-year-old men and women. Arch. Osteoporos 2018, 13, 9. [Google Scholar] [CrossRef] [Green Version]
- Hanzlik, R.P.; Fowler, S.C.; Fisher, D.H. Relative bioavailability of calcium from calcium formate, calcium citrate, and calcium carbonate. J. Pharmacol. Exp. Ther. 2005, 313, 1217–1222. [Google Scholar] [CrossRef]
- Cormick, G.; Belizán, J.M. Calcium Intake and Health. Nutrients 2019, 11, 1606. [Google Scholar] [CrossRef] [Green Version]
- Khazai, N.; Judd, S.E.; Tangpricha, V. Calcium and vitamin D: Skeletal and extraskeletal health. Curr. Rheumatol. Rep. 2008, 10, 110–117. [Google Scholar] [CrossRef] [Green Version]
- Wasilewski, G.B.; Vervloet, M.G.; Schurgers, L.J. The Bone-Vasculature Axis: Calcium Supplementation and the Role of Vitamin K. Front. Cardiovasc. Med. 2019, 6, 6. [Google Scholar] [CrossRef]
- Calvez, J.; Poupin, N.; Chesneau, C.; Lassale, C.; Tomé, D. Protein intake, calcium balance and health consequences. Eur. J. Clin. Nutr. 2012, 66, 281–295. [Google Scholar] [CrossRef] [Green Version]
- Alharbi, O.; El-Sohemy, A. Lactose Intolerance (LCT-13910C>T) Genotype Is Associated with Plasma 25-Hydroxyvitamin D Concentrations in Caucasians: A Mendelian Randomization Study. J. Nutr. 2017, 147, 1063–1069. [Google Scholar] [CrossRef] [Green Version]
- Itkonen, S.T.; Erkkola, M.; Lamberg-Allardt, C.J.E. Vitamin D Fortification of Fluid Milk Products and Their Contribution to Vitamin D Intake and Vitamin D Status in Observational Studies—A Review. Nutrients 2018, 10, 1054. [Google Scholar] [CrossRef] [Green Version]
- Polzonetti, V.; Pucciarelli, S.; Vincenzetti, S.; Polidori, P. Dietary Intake of Vitamin D from Dairy Products Reduces the Risk of Osteoporosis. Nutrients 2020, 12, 1743. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.Q.; Xu, J.Y.; Han, S.F.; Zhang, Z.L.; Zhao, Y.Y.; Szeto, I.M. Dairy consumption and risk of cardiovascular disease: An updated meta-analysis of prospective cohort studies. Asia Pac. J. Clin. Nutr. 2015, 24, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Dehghan, M.; Mente, A.; Rangarajan, S.; Sheridan, P.; Mohan, V.; Iqbal, R.; Gupta, R.; Lear, S.; Wentzel-Viljoen, E.; Avezum, A.; et al. Association of dairy intake with cardiovascular disease and mortality in 21 countries from five continents (PURE): A prospective cohort study. Lancet 2018, 392, 2288–2297. [Google Scholar] [CrossRef]
- Alexander, D.D.; Bylsma, L.C.; Vargas, A.J.; Cohen, S.S.; Doucette, A.; Mohamed, M.; Irvin, S.R.; Miller, P.E.; Watson, H.; Fryzek, J.P. Dairy consumption and CVD: A systematic review and meta-analysis. Br. J. Nutr. 2016, 115, 737–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamarche, B.; Givens, D.I.; Soedamah-Muthu, S.; Krauss, R.M.; Jakobsen, M.U.; Bischoff-Ferrari, H.A.; Pan, A.; Després, J.P. Does Milk Consumption Contribute to Cardiometabolic Health and Overall Diet Quality? Can. J. Cardiol. 2016, 32, 1026–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Givens, D.I.; Astrup, A.; Bakker, S.J.L.; Goossens, G.H.; Kratz, M.; Marette, A.; Pijl, H.; Soedamah-Muthu, S.S. The Impact of Dairy Products in the Development of Type 2 Diabetes: Where Does the Evidence Stand in 2019? Adv. Nutr. 2019, 10, 1066–1075. [Google Scholar] [CrossRef] [Green Version]
- Brown-Riggs, C. Nutrition and Health Disparities: The Role of Dairy in Improving Minority Health Outcomes. Int. J. Environ. Res. Public Health 2015, 13, 28. [Google Scholar] [CrossRef] [Green Version]
- Gille, D.; Schmid, A.; Walther, B.; Vergères, G. Fermented Food and Non-Communicable Chronic Diseases: A Review. Nutrients 2018, 10, 448. [Google Scholar] [CrossRef] [Green Version]
- Szilagyi, A. Complex Interactions of Obesity, Dairy Food Intake and Genetics of Lactase. J. Obes. Chronic Dis. 2018, 2. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Sundquist, J.; Sundquist, K. Lactose intolerance and risk of lung, breast and ovarian cancers: Aetiological clues from a population-based study in Sweden. Br. J. Cancer 2015, 112, 149–152. [Google Scholar] [CrossRef] [Green Version]
- Grosso, G.; Bella, F.; Godos, J.; Sciacca, S.; Del Rio, D.; Ray, S.; Galvano, F.; Giovannucci, E.L. Possible role of diet in cancer: Systematic review and multiple meta-analyses of dietary patterns, lifestyle factors, and cancer risk. Nutr. Rev. 2017, 75, 405–419. [Google Scholar] [CrossRef]
- Aune, D.; Lau, R.; Chan, D.S.M.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Dairy products and colorectal cancer risk: A systematic review and meta-analysis of cohort studies. Ann. Oncol. 2012, 23, 37–45. [Google Scholar] [CrossRef]
- Murphy, N.; Norat, T.; Ferrari, P.; Jenab, M.; Bueno-de-Mesquita, B.; Skeie, G.; Olsen, A.; Tjønneland, A.; Dahm, C.C.; Overvad, K.; et al. Consumption of dairy products and colorectal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC). PLoS ONE 2013, 8, e72715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Hu, W.; Liu, W.X.; Zhao, L.Y.; Huang, D.; Liu, X.D.; Chan, H.; Zhang, Y.; Zeng, J.D.; Coker, O.O.; et al. Streptococcus thermophilus Inhibits Colorectal Tumorigenesis Through Secreting β-Galactosidase. Gastroenterology 2021, 160, 1179–1193.e1114. [Google Scholar] [CrossRef]
- Kaluza, J.; Komatsu, S.; Lauriola, M.; Harris, H.R.; Bergkvist, L.; Michaëlsson, K.; Wolk, A. Long-term consumption of non-fermented and fermented dairy products and risk of breast cancer by estrogen receptor status—Population-based prospective cohort study. Clin. Nutr. 2020, 40, 1966–1973. [Google Scholar] [CrossRef]
- Zhang, K.; Dai, H.; Liang, W.; Zhang, L.; Deng, Z. Fermented dairy foods intake and risk of cancer. Int. J. Cancer 2019, 144, 2099–2108. [Google Scholar] [CrossRef]
- Ito, M.; Kimura, M. Influence of Lactose on Faecal Microflora in Lactose Maldigestors. Microb. Ecol. Health Dis. 1993, 6, 73–76. [Google Scholar] [CrossRef]
- He, T.; Priebe, M.G.; Zhong, Y.; Huang, C.; Harmsen, H.J.; Raangs, G.C.; Antoine, J.M.; Welling, G.W.; Vonk, R.J. Effects of yogurt and bifidobacteria supplementation on the colonic microbiota in lactose-intolerant subjects. J. Appl. Microbiol. 2008, 104, 595–604. [Google Scholar] [CrossRef]
- Li, X.; Yin, J.; Zhu, Y.; Wang, X.; Hu, X.; Bao, W.; Huang, Y.; Chen, L.; Chen, S.; Yang, W.; et al. Effects of Whole Milk Supplementation on Gut Microbiota and Cardiometabolic Biomarkers in Subjects with and without Lactose Malabsorption. Nutrients 2018, 10, 1403. [Google Scholar] [CrossRef] [Green Version]
- Szilagyi, A.; Shrier, I.; Heilpern, D.; Je, J.; Park, S.; Chong, G.; Lalonde, C.; Cote, L.F.; Lee, B. Differential impact of lactose/lactase phenotype on colonic microflora. Can. J. Gastroenterol. 2010, 24, 373–379. [Google Scholar] [CrossRef]
- Szilagyi, A. Redefining lactose as a conditional prebiotic. Can. J. Gastroenterol. 2004, 18, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Hertzler, S.R.; Savaiano, D.A. Colonic adaptation to daily lactose feeding in lactose maldigesters reduces lactose intolerance. Am. J. Clin. Nutr. 1996, 64, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.O.; Semenya, J.G.; Buchowski, M.S.; Enwonwu, C.O.; Scrimshaw, N.S. Adaptation of lactose maldigesters to continued milk intakes. Am. J. Clin. Nutr. 1993, 58, 879–881. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R.; De Caterina, R.; Görman, U.; Allayee, H.; Kohlmeier, M.; Prasad, C.; Choi, M.S.; Curi, R.; de Luis, D.A.; Gil, Á.; et al. Guide and Position of the International Society of Nutrigenetics/Nutrigenomics on Personalised Nutrition: Part 1—Fields of Precision Nutrition. J. Nutr. Nutr. 2016, 9, 12–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aruoma, O.I.; Hausman-Cohen, S.; Pizano, J.; Schmidt, M.A.; Minich, D.M.; Joffe, Y.; Brandhorst, S.; Evans, S.J.; Brady, D.M. Personalized Nutrition: Translating the Science of NutriGenomics Into Practice: Proceedings From the 2018 American College of Nutrition Meeting. J. Am. Coll. Nutr. 2019, 38, 287–301. [Google Scholar] [CrossRef]
- Anguita-Ruiz, A.; Aguilera, C.M.; Gil, Á. Genetics of Lactose Intolerance: An Updated Review and Online Interactive World Maps of Phenotype and Genotype Frequencies. Nutrients 2020, 12, 2689. [Google Scholar] [CrossRef]
- Almon, R.; Engfeldt, P.; Tysk, C.; Sjöström, M.; Nilsson, T.K. Prevalence and trends in adult-type hypolactasia in different age cohorts in Central Sweden diagnosed by genotyping for the adult-type hypolactasia-linked LCT -13910C > T mutation. Scand. J. Gastroenterol. 2007, 42, 165–170. [Google Scholar] [CrossRef]
- Lember, M.; Torniainen, S.; Kull, M.; Kallikorm, R.; Saadla, P.; Rajasalu, T.; Komu, H.; Järvelä, I. Lactase non-persistence and milk consumption in Estonia. World J. Gastroenterol. 2006, 12, 7329–7331. [Google Scholar] [CrossRef]
- Imtiaz, F.; Savilahti, E.; Sarnesto, A.; Trabzuni, D.; Al-Kahtani, K.; Kagevi, I.; Rashed, M.S.; Meyer, B.F.; Järvelä, I. The T/G 13915 variant upstream of the lactase gene (LCT) is the founder allele of lactase persistence in an urban Saudi population. J. Med. Genet. 2007, 44, e89. [Google Scholar] [CrossRef] [Green Version]
- Floris, M.; Cano, A.; Porru, L.; Addis, R.; Cambedda, A.; Idda, M.L.; Steri, M.; Ventura, C.; Maioli, M. Direct-to-Consumer Nutrigenetics Testing: An Overview. Nutrients 2020, 12, 566. [Google Scholar] [CrossRef] [Green Version]
- Guasch-Ferré, M.; Dashti, H.S.; Merino, J. Nutritional Genomics and Direct-to-Consumer Genetic Testing: An Overview. Adv. Nutr. 2018, 9, 128–135. [Google Scholar] [CrossRef]
- Marietta, C.; McGuire, A.L. Currents in contemporary ethics. Direct-to-consumer genetic testing: Is it the practice of medicine? J. Law Med. Ethics 2009, 37, 369–374. [Google Scholar] [CrossRef]
- De, S.; Pietilä, A.M.; Iso-Touru, T.; Hopia, A.; Tahvonen, R.; Vähäkangas, K. Information Provided to Consumers about Direct-to-Consumer Nutrigenetic Testing. Public Health Genom. 2019, 22, 162–173. [Google Scholar] [CrossRef]
- Nicklas, T.A.; Qu, H.; Hughes, S.O.; He, M.; Wagner, S.E.; Foushee, H.R.; Shewchuk, R.M. Self-perceived lactose intolerance results in lower intakes of calcium and dairy foods and is associated with hypertension and diabetes in adults. Am. J. Clin. Nutr. 2011, 94, 191–198. [Google Scholar] [CrossRef] [Green Version]
- Kalokairinou, L.; Howard, H.; Slokenberga, S.; Fisher, E.; Thoeni, M.; Hartlev, M.; Hellemondt, R.; Juškevičius, J.; Kapelenska-Pregowska, J.; Kovac, P.; et al. Legislation of direct-to-consumer genetic testing in Europe: A fragmented regulatory landscape. J. Community Genet. 2018, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Tandy-Connor, S.; Guiltinan, J.; Krempely, K.; LaDuca, H.; Reineke, P.; Gutierrez, S.; Gray, P.; Tippin Davis, B. False-positive results released by direct-to-consumer genetic tests highlight the importance of clinical confirmation testing for appropriate patient care. Genet. Med. 2018, 20, 1515–1521. [Google Scholar] [CrossRef] [Green Version]
- Labrie, V.; Buske, O.J.; Oh, E.; Jeremian, R.; Ptak, C.; Gasiūnas, G.; Maleckas, A.; Petereit, R.; Žvirbliene, A.; Adamonis, K.; et al. Lactase nonpersistence is directed by DNA-variation-dependent epigenetic aging. Nat. Struct. Mol. Biol. 2016, 23, 566–573. [Google Scholar] [CrossRef] [Green Version]
- Leseva, M.N.; Grand, R.J.; Klett, H.; Boerries, M.; Busch, H.; Binder, A.M.; Michels, K.B. Differences in DNA Methylation and Functional Expression in Lactase Persistent and Non-persistent Individuals. Sci. Rep. 2018, 8, 5649. [Google Scholar] [CrossRef]
- Fukushima, A.; Goda, T.; Motohashi, Y.; Sakuma, K. The specific expression patterns of lactase, sucrase and calbindin-D9k in weaning rats are regulated at the transcriptional level. J. Nutr. Sci. Vitam. 2004, 50, 265–271. [Google Scholar] [CrossRef] [Green Version]
- Motohashi, Y.; Fukushima, A.; Kondo, T.; Sakuma, K. Lactase decline in weaning rats is regulated at the transcriptional level and not caused by termination of milk ingestion. J. Nutr. 1997, 127, 1737–1743. [Google Scholar] [CrossRef] [Green Version]
- Rossi, M.; Maiuri, L.; Fusco, M.I.; Salvati, V.M.; Fuccio, A.; Auricchio, S.; Mantei, N.; Zecca, L.; Gloor, S.M.; Semenza, G. Lactase persistence versus decline in human adults: Multifactorial events are involved in down-regulation after weaning. Gastroenterology 1997, 112, 1506–1514. [Google Scholar] [CrossRef]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Nair, G.B. Homeostasis and dysbiosis of the gut microbiome in health and disease. J. Biosci. 2019, 44, 1–8. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Holtug, K.; Clausen, M.R.; Hove, H.; Christiansen, J.; Mortensen, P.B. The colon in carbohydrate malabsorption: Short-chain fatty acids, pH, and osmotic diarrhoea. Scand. J. Gastroenterol. 1992, 27, 545–552. [Google Scholar] [CrossRef]
- Clausen, M.R.; Mortensen, P.B. Lactulose, disaccharides and colonic flora. Clinical consequences. Drugs 1997, 53, 930–942. [Google Scholar] [CrossRef]
- Zhong, Y.; Priebe, M.G.; Vonk, R.J.; Huang, C.Y.; Antoine, J.M.; He, T.; Harmsen, H.J.; Welling, G.W. The role of colonic microbiota in lactose intolerance. Dig. Dis. Sci. 2004, 49, 78–83. [Google Scholar] [CrossRef]
- Azcarate-Peril, M.A.; Ritter, A.J.; Savaiano, D.; Monteagudo-Mera, A.; Anderson, C.; Magness, S.T.; Klaenhammer, T.R. Impact of short-chain galactooligosaccharides on the gut microbiome of lactose-intolerant individuals. Proc. Natl. Acad. Sci. USA 2017, 114, E367–E375. [Google Scholar] [CrossRef] [Green Version]
- Arnold, J.W.; Simpson, J.B.; Roach, J.; Bruno-Barcena, J.M.; Azcarate-Peril, M.A. Prebiotics for Lactose Intolerance: Variability in Galacto-Oligosaccharide Utilization by Intestinal Lactobacillus rhamnosus. Nutrients 2018, 10, 1517. [Google Scholar] [CrossRef] [Green Version]
- Jiang, T.; Mustapha, A.; Savaiano, D.A. Improvement of lactose digestion in humans by ingestion of unfermented milk containing Bifidobacterium longum. J. Dairy Sci. 1996, 79, 750–757. [Google Scholar] [CrossRef]
- Savaiano, D.A.; Ritter, A.J.; Klaenhammer, T.R.; James, G.M.; Longcore, A.T.; Chandler, J.R.; Walker, W.A.; Foyt, H.L. Improving lactose digestion and symptoms of lactose intolerance with a novel galacto-oligosaccharide (RP-G28): A randomized, double-blind clinical trial. Nutr. J. 2013, 12, 160. [Google Scholar] [CrossRef] [Green Version]
- Leis, R.; de Castro, M.J.; de Lamas, C.; Picáns, R.; Couce, M.L. Effects of Prebiotic and Probiotic Supplementation on Lactase Deficiency and Lactose Intolerance: A Systematic Review of Controlled Trials. Nutrients 2020, 12, 1487. [Google Scholar] [CrossRef]
- Oak, S.J.; Jha, R. The effects of probiotics in lactose intolerance: A systematic review. Crit. Rev. Food Sci. Nutr. 2019, 59, 1675–1683. [Google Scholar] [CrossRef]
- Gingold-Belfer, R.; Levy, S.; Layfer, O.; Pakanaev, L.; Niv, Y.; Dickman, R.; Perets, T.T. Use of a Novel Probiotic Formulation to Alleviate Lactose Intolerance Symptoms-a Pilot Study. Probiotics Antimicrob. Proteins 2020, 12, 112–118. [Google Scholar] [CrossRef]
- Ulaszewska, M.M.; Weinert, C.H.; Trimigno, A.; Portmann, R.; Andres Lacueva, C.; Badertscher, R.; Brennan, L.; Brunius, C.; Bub, A.; Capozzi, F.; et al. Nutrimetabolomics: An Integrative Action for Metabolomic Analyses in Human Nutritional Studies. Mol. Nutr. Food Res. 2019, 63, e1800384. [Google Scholar] [CrossRef]
- O’Gorman, A.; Gibbons, H.; Brennan, L. Metabolomics in the identification of biomarkers of dietary intake. Comput. Struct. Biotechnol. J. 2013, 4, e201301004. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, G.; Burton, K.J.; Rosikiewicz, M.; Freiburghaus, C.; von Ah, U.; Münger, L.H.; Pralong, F.P.; Vionnet, N.; Greub, G.; Badertscher, R.; et al. Blood lactose after dairy product intake in healthy men. Br. J. Nutr. 2017, 118, 1070–1077. [Google Scholar] [CrossRef] [Green Version]
- Koulman, A.; Volmer, D.A. Perspectives for Metabolomics in Human Nutrition: An Overview. Nutr. Bull. 2008, 33, 324–330. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, G.; Burnand, D.; Münger, L.H.; Pralong, F.P.; Vionnet, N.; Portmann, R.; Vergères, G. Identification of Milk and Cheese Intake Biomarkers in Healthy Adults Reveals High Interindividual Variability of Lewis System–Related Oligosaccharides. J. Nutr. 2020, 150, 1058–1067. [Google Scholar] [CrossRef] [Green Version]
- Vionnet, N.; Münger, L.H.; Freiburghaus, C.; Burton, K.J.; Pimentel, G.; Pralong, F.P.; Badertscher, R.; Vergères, G. Assessment of lactase activity in humans by measurement of galactitol and galactonate in serum and urine after milk intake. Am. J. Clin. Nutr. 2019, 109, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Cold Spring Harb Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tebani, A.; Bekri, S. Paving the Way to Precision Nutrition Through Metabolomics. Front. Nutr. 2019, 6, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimentel, G.; Burton, K.J.; von Ah, U.; Butikofer, U.; Pralong, F.P.; Vionnet, N.; Portmann, R.; Vergeres, G. Metabolic Footprinting of Fermented Milk Consumption in Serum of Healthy Men. J. Nutr. 2018, 148, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Kalache, A.; de Hoogh, A.I.; Howlett, S.E.; Kennedy, B.; Eggersdorfer, M.; Marsman, D.S.; Shao, A.; Griffiths, J.C. Nutrition interventions for healthy ageing across the lifespan: A conference report. Eur. J. Nutr. 2019, 58, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Authority, E.F.S. Dietary Reference Values for nutrients Summary report. EFSA Supporting Publ. 2017, 14, e15121E. [Google Scholar] [CrossRef] [Green Version]
- Mathers, J.C. Paving the way to better population health through personalised nutrition. EFSA J. 2019, 17, e170713. [Google Scholar] [CrossRef]
- Dwyer, J. Dietary Reference Intakes (DRIs): Concepts and Implementation. In Encyclopedia of Gastroenterology; Johnson, L.R., Ed.; Elsevier: New York, NY, USA, 2004; pp. 613–623. [Google Scholar] [CrossRef]
- Huth, P.J.; DiRienzo, D.B.; Miller, G.D. Major Scientific Advances with Dairy Foods in Nutrition and Health. J. Dairy Sci. 2006, 89, 1207–1221. [Google Scholar] [CrossRef]
- Department of Health. Dietary Reference Values: A Guide; H.M. Stationery Office: London, UK, 1991.
- Mutch, D.M.; Wahli, W.; Williamson, G. Nutrigenomics and nutrigenetics: The emerging faces of nutrition. FASEB J. 2005, 19, 1602–1616. [Google Scholar] [CrossRef]
- Gaboon, N.E.A. Nutritional genomics and personalized diet. Egypt. J. Med. Hum. Genet. 2011, 12, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Gorduza, E.V.; Indrei, L.L.; Gorduza, V.M. Nutrigenomics in postgenomic era. Rev. Med. Chir Soc. Med. Nat. Iasi 2008, 112, 152–164. [Google Scholar]
- Beckett, E.; Jones, P.; Veysey, M.; Lucock, M. Nutrigenetics—Personalized Nutrition in the Genetic Age. Explor. Res. Hypothesis Med. 2017, 2, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Kanter, M.; Desrosiers, A. Personalized Wellness Past and Future: Will the Science and Technology Coevolve? Nutr. Today 2019, 54, 174–181. [Google Scholar] [CrossRef]
- Celis-Morales, C.; Marsaux, C.F.; Livingstone, K.M.; Navas-Carretero, S.; San-Cristobal, R.; Fallaize, R.; Macready, A.L.; O’Donovan, C.; Woolhead, C.; Forster, H.; et al. Can genetic-based advice help you lose weight? Findings from the Food4Me European randomized controlled trial. Am. J. Clin. Nutr. 2017, 105, 1204–1213. [Google Scholar] [CrossRef] [Green Version]
- Ghvanidze, S.; Velikova, N.; Dodd, T.; Wilna, O.-T. Consumers’ environmental and ethical consciousness and the use of the related food products information: The role of perceived consumer effectiveness. Appetite 2016, 107, 311–322. [Google Scholar] [CrossRef]
- Sanchez-Sabate, R.; Sabaté, J. Consumer Attitudes Towards Environmental Concerns of Meat Consumption: A Systematic Review. Int. J. Environ. Res. Public Health 2019, 16, 1220. [Google Scholar] [CrossRef] [Green Version]
- Roosen, J.; Bruhn, M.; Mecking, R.A.; Drescher, L.S. Consumer demand for personalized nutrition and functional food. Int. J. Vitam. Nutr. Res. 2008, 78, 269–274. [Google Scholar] [CrossRef]
- Vallée Marcotte, B.; Cormier, H.; Garneau, V.; Robitaille, J.; Desroches, S.; Vohl, M.C. Current knowledge and interest of French Canadians regarding nutrigenetics. Genes Nutr. 2019, 14, 5. [Google Scholar] [CrossRef]
- Chaudhary, N.; Kumar, V.; Sangwan, P.; Pant, N.C.; Saxena, A.; Joshi, S.; Yadav, A.N. Personalized Nutrition and -Omics. Compr. Foodomics 2021, 495–507. [Google Scholar] [CrossRef]
- Ghosh, D. Future perspectives of nutrigenomics foods: Benefits vs. risks. Indian J. Biochem. Biophys. 2009, 46, 31–36. [Google Scholar]
- Chetty, R.; Stepner, M.; Abraham, S.; Lin, S.; Scuderi, B.; Turner, N.; Bergeron, A.; Cutler, D. The Association Between Income and Life Expectancy in the United States, 2001–2014. JAMA 2016, 315, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.H.; Anthony, J.C.; Carvajal, R.; Chae, L.; Khoo, C.S.H.; Latulippe, M.E.; Matusheski, N.V.; McClung, H.L.; Rozga, M.; Schmid, C.H.; et al. Perspective: Guiding Principles for the Implementation of Personalized Nutrition Approaches That Benefit Health and Function. Adv. Nutr. 2019, 11, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Sorrenti, V.; Caudullo, G.; Lucignano, F.; Fortinguerra, S.; Zusso, M.; Giusti, P.; Buriani, A. Chapter Eighteen—Personalized sports nutrition: Role of nutrients in athletic performance. In Sports, Exercise, and Nutritional Genomics; Barh, D., Ahmetov, I.I., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 411–431. [Google Scholar] [CrossRef]
- Ong, M.L.; Lin, X.; Holbrook, J.D. Measuring epigenetics as the mediator of gene/environment interactions in DOHaD. J. Dev. Orig. Health Dis. 2015, 6, 10–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picó, C.; Serra, F.; Rodríguez, A.M.; Keijer, J.; Palou, A. Biomarkers of Nutrition and Health: New Tools for New Approaches. Nutrients 2019, 11, 1092. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.X. Identification of Metabolic Biomarkers for Personalized Nutrition. Lifestyle Genom. 2012, 5, 1–2. [Google Scholar] [CrossRef]
- Shrestha, A.; Barnett, M.P.G.; Perry, J.K.; Cameron-Smith, D.; Milan, A.M. Evaluation of breath, plasma, and urinary markers of lactose malabsorption to diagnose lactase non-persistence following lactose or milk ingestion. BMC Gastroenterol. 2020, 20, 204. [Google Scholar] [CrossRef]
- Lillie, E.O.; Patay, B.; Diamant, J.; Issell, B.; Topol, E.J.; Schork, N.J. The n-of-1 clinical trial: The ultimate strategy for individualizing medicine? Pers. Med. 2011, 8, 161–173. [Google Scholar] [CrossRef] [Green Version]
- Fassio, F.; Facioni, M.S.; Guagnini, F. Lactose Maldigestion, Malabsorption, and Intolerance: A Comprehensive Review with a Focus on Current Management and Future Perspectives. Nutrients 2018, 10, 1599. [Google Scholar] [CrossRef] [Green Version]
- EFSA Panel on Dietetic Products, Nutrition and Allergies. Scientific Opinion on lactose thresholds in lactose intolerance and galactosaemia. EFSA J. 2010, 8, 1777. [Google Scholar] [CrossRef]
- Röttger-Wirtz, S.; De Boer, A. Personalised Nutrition: The EU’s Fragmented Legal Landscape and the Overlooked Implications of EU Food Law. Eur. J. Risk Regul. 2020, 1–24. [Google Scholar] [CrossRef]
- Nordström, K.; Goossens, J. Personalized Nutrition and Social Justice: Ethical Considerations Within Four Future Scenarios Applying the Perspective of Nussbaum’s Capabilities Approach. J. Agric. Environ. Ethics 2016, 29, 5–22. [Google Scholar] [CrossRef]
- Wiley, A.S. “Drink Milk for Fitness”: The Cultural Politics of Human Biological Variation and Milk Consumption in the United States. Am. Anthropol. 2004, 106, 506–517. [Google Scholar] [CrossRef]
- Casellas, F.; Aparici, A.; Pérez, M.J.; Rodríguez, P. Perception of lactose intolerance impairs health-related quality of life. Eur. J. Clin. Nutr. 2016, 70, 1068–1072. [Google Scholar] [CrossRef]
- Ledochowski, M.; Sperner-Unterweger, B.; Fuchs, D. Lactose malabsorption is associated with early signs of mental depression in females: A preliminary report. Dig. Dis. Sci. 1998, 43, 2513–2517. [Google Scholar] [CrossRef]
- Enko, D.; Meinitzer, A.; Brandmayr, W.; Halwachs-Baumann, G.; Schnedl, W.J.; Kriegshäuser, G. Association between increased plasma levels of homocysteine and depression observed in individuals with primary lactose malabsorption. PLoS ONE 2018, 13, e0202567. [Google Scholar] [CrossRef] [Green Version]
- Meijboom, F.L.B.; Verweij, M.F.; Brom, F.W.A. You Eat What You Are: Moral Dimensions of Diets Tailored to One’s Genes. J. Agric. Environ. Ethics 2003, 16, 557–568. [Google Scholar] [CrossRef]
- Mayes, C. Medicalization of Eating and Feeding. In Encyclopedia of Food and Agricultural Ethics; Thompson, P.B., Kaplan, D.M., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 1–8. [Google Scholar] [CrossRef] [Green Version]
| 1 | Phenotype expressed by the continued activity of the lactase enzyme throughout adulthood. |

| Term | Definition |
|---|---|
| Lactose malabsorption | Failure to digest/absorb lactose due to primary or secondary lactase deficiency. |
| Lactose intolerance (LI) | Clinical syndrome in which the ingestion of lactose causes typical gastrointestinal symptoms such as diarrhea, bloating, flatulence, nausea, abdominal pain, cramps. |
| Self-reported LI | Individuals who perceive themselves as being LI without medical diagnosis. |
| Lactase deficiency | Lack or absence of intestinal lactase enzyme activity. |
| Congenital lactase deficiency | Rare genetic disorder in which lactase is already absent at birth. |
| Primary lactase deficiency | Progressive decline of lactase enzyme activity with age. |
| Secondary lactase deficiency | Reversible condition caused by illness or injury of the small intestine and resulting in deficiency of intestinal lactase enzyme activity. |
| Lactase non-persistence (LNP) | Most common phenotype associated with lactase gene expression worldwide. Characterized by lactase activity decline during early childhood. |
| Lactase persistence (LP) | Phenotype expressed by the continued activity of the lactase enzyme throughout adulthood. |
| Country or Population | Alleles | Frequency (%) | Reference |
|---|---|---|---|
| Sweden | LCT-13910C/T | 73.7 | [118] |
| Estonia | LCT-13910C/T | 51.4 | [119] |
| Saudi Arabia | LCT-13915T/G | 59.4 | [120] |
| Jordan | LCT-13915T/G | 39.1 | [26] |
| Tanzania | LCT-14010G/C | 31.9 | [11] |
| Kenya | LCT-14010G/C | 27.6 | [19] |
| Ethiopia (Afar) | LCT-13907C/G | 20.0 | [26] |
| Sudan (Afro-Asiatic Beja) | LCT-13907C/G | 20.6 | [11] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porzi, M.; Burton-Pimentel, K.J.; Walther, B.; Vergères, G. Development of Personalized Nutrition: Applications in Lactose Intolerance Diagnosis and Management. Nutrients 2021, 13, 1503. https://doi.org/10.3390/nu13051503
Porzi M, Burton-Pimentel KJ, Walther B, Vergères G. Development of Personalized Nutrition: Applications in Lactose Intolerance Diagnosis and Management. Nutrients. 2021; 13(5):1503. https://doi.org/10.3390/nu13051503
Chicago/Turabian StylePorzi, Millie, Kathryn J. Burton-Pimentel, Barbara Walther, and Guy Vergères. 2021. "Development of Personalized Nutrition: Applications in Lactose Intolerance Diagnosis and Management" Nutrients 13, no. 5: 1503. https://doi.org/10.3390/nu13051503