Dietary Curdlan Enhances Bifidobacteria and Reduces Intestinal Inflammation in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
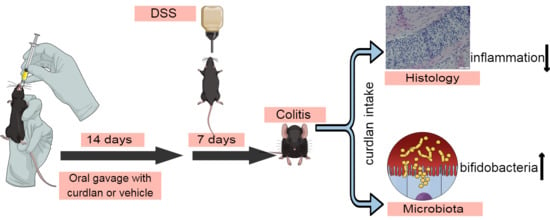
2.2. DSS-Induced Colitis Experiment
2.3. Histology
2.4. Isolation and Culture of Peritoneal Macrophages (PM) in Ex Vivo Set Up
2.5. mRNA Extraction, cDNA Synthesis and qPCR Analysis
2.6. Measurement of Cytokine Protein Concentrations
2.7. Human i-Screen
Quantification of Total DNA (qPCR)
2.8. DNA Isolation for Microbiome and Mycobiome Sequencing of Mice Colon Contents
2.9. Bacterial Profiling from Mouse Colon Contents and Human i-Screen-Derived DNA Amplicons
2.10. Fungal Profiling from Mouse Colon Contents
2.11. Statistical Analyses
3. Results
3.1. Curdlan Intake Reduces Signs of Inflammation in Acute DSS-Induced Colitis in Mice
3.2. Two Weeks of Dietary Curdlan Reduces LPS Responses in Peritoneal Macrophages
3.3. Mouse Colonic Microbial Diversity Is Affected by Dietary Curdlan
3.4. Bifidobacterium Abundantly Present after Curdlan Feeding in Mice
3.5. Curdlan Additionally, Affects Human Microbial Composition
3.6. Relative Abundance of Bifidobacterium Choerinum in Mice and Not Human
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nakashima, A.; Yamada, K.; Iwata, O.; Sugimoto, R.; Atsuji, K.; Ogawa, T.; Ishibashi-Ohgo, N.; Suzuki, K. β-Glucan in Foods and Its Physiological Functions. J. Nutr. Sci. Vitaminol. 2018, 64, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Freitas, F.; Alves, V.D.; Reis, M.A. Advances in bacterial exopolysaccharides: From production to biotechnological applications. Trends Biotechnol. 2011, 29, 388–398. [Google Scholar] [CrossRef]
- Zhang, R.; Edgar, K.J. Properties, Chemistry, and Applications of the Bioactive Polysaccharide Curdlan. Biomacromolecules 2014, 15, 1079–1096. [Google Scholar] [CrossRef]
- Lee, K.B.; Bae, J.H.; Kim, J.S.; Yoo, Y.C.; Kim, B.S.; Kwak, S.T.; Kim, Y.S. Anticoagulant activity of sulfoalkyl derivatives of curdlan. Arch. Pharmacol Res. 2001, 24, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Jagodzinski, P.P.; Wiaderkiewicz, R.; Kurzawski, G.; Kloczewiak, M.; Nakashima, H.; Hyjek, E.; Yamamoto, N.; Uryu, T.; Kaneko, Y.; Posner, M.R.; et al. Mechanism of the Inhibitory Effect of Curdlan Sulfate on HIV-1 Infection in Vitro. Virology 1994, 202, 735–745. [Google Scholar] [CrossRef]
- Spicer, E.; Goldenthal, E.; Ikeda, T. A toxicological assessment of curdlan. Food Chem. Toxicol. 1999, 37, 455–479. [Google Scholar] [CrossRef]
- Kataoka, K.; Tatsushi, M.; Yamazaki, S.; Takeshige, K. Activation of macrophages by linear (1right-arrow3)-beta-D-glucans. Impliations for the recognition of fungi by innate immunity. J. Biol. Chem. 2002, 277, 36825–36831. [Google Scholar] [CrossRef] [Green Version]
- Steele, C.; Marrero, L.; Swain, S.D.; Harmsen, A.G.; Zheng, M.; Brown, G.D.; Gordon, S.; Shellito, J.E.; Kolls, J.K. Alveolar Macrophage–mediated Killing of Pneumocystis carinii f. sp. muris Involves Molecular Recognition by the Dectin-1 β-Glucan Receptor. J. Exp. Med. 2003, 198, 1677–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Zhao, Y. The Biological Role of Dectin-1 in Immune Response. Int. Rev. Immunol. 2007, 26, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.R.; Brown, G.D. The role of Dectin-1 in the host defence against fungal infections. Curr. Opin. Microbiol. 2011, 14, 392–399. [Google Scholar] [CrossRef]
- Marakalala, M.J.; Kerrigan, A.M.; Brown, G.D. Dectin-1: A role in antifungal defense and consequences of genetic polymorphisms in humans. Mamm. Genome 2011, 22, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Rahabi, M.; Jacquemin, G.; Prat, M.; Meunier, E.; Alaeddine, M.; Bertrand, B.; Lefèvre, L.; Benmoussa, K.; Batigne, P.; Aubouy, A.; et al. Divergent Roles for Macrophage C-type Lectin Receptors, Dectin-1 and Mannose Receptors, in the Intestinal Inflammatory Response. Cell Rep. 2020, 30, 4386–4398. [Google Scholar] [CrossRef] [PubMed]
- Bădulescu, M.-M.; Apetrei, N.S.; Lupu, A.-R.; Cremer, L.; Szegli, G.; Moscovici, M.; Mocanu, G.; Mihai, D.; Călugăru, A. Curdlan derivatives able to enhance cytostatic drugs activity on tumor cells. Roum. Arch. Microbiol. Immunol. 2010, 68, 201. [Google Scholar]
- Rui, K.; Tian, J.; Tang, X.; Ma, J.; Xu, P.; Tian, X.; Wang, Y.; Xu, H.; Lu, L.; Wang, S. Curdlan blocks the immune suppression by myeloid-derived suppressor cells and reduces tumor burden. Immunol. Res. 2016, 64, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Negi, S.; Pahari, S.; Das, D.K.; Khan, N.; Agrewala, J.N. Curdlan Limits Mycobacterium tuberculosis Survival Through STAT-1 Regulated Nitric Oxide Production. Front. Microbiol. 2019, 10, 1173. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Liu, J.; Yan, Q.; You, X.; Yang, S.; Jiang, Z. In vitro digestibility and prebiotic potential of curdlan (1 → 3)-β- d -glucan oligosaccharides in Lactobacillus species. Carbohydr. Polym. 2018, 188, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Rajulapati, V.; Goyal, A. In vitro prebiotic potential, digestibility and biocompatibility properties of laminari-oligosaccharides produced from curdlan by β-1,3-endoglucanase from Clostridium thermocellum. 3 Biotech 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Verma, D.K.; Niamah, A.K.; Patel, A.R.; Thakur, M.; Sandhu, K.S.; Chávez-González, M.L.; Shah, N.; Aguilar, C.N. Chemistry and microbial sources of curdlan with potential application and safety regulations as prebiotic in food and health. Food Res. Int. 2020, 133, 109136. [Google Scholar] [CrossRef]
- Shimizu, J.; Tsuchihashi, N.; Kudoh, K.; Wada, M.; Takita, T.; Innami, S. Dietary Curdlan Increases Proliferation of Bifidobacteria in The Cecum of Rats. Biosci. Biotechnol. Biochem. 2001, 65, 466–469. [Google Scholar] [CrossRef]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willing, B.; Halfvarson, J.; Dicksved, J.; Rosenquist, M.; Jarnerot, G.; Engstrand, L.; Tysk, C.; Jansson, J.K. Twin Studies Reveal Specific Imbalances in the Mucosaassociated Microbiota of Patients with Ileal Crohn’s Disease. Inflamm. Bowel Dis. 2008, 15, 653–660. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henson, M.A.; Phalak, P. Microbiota dysbiosis in inflammatory bowel diseases: In silico investigation of the oxygen hypothesis. BMC Syst. Biol. 2017, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, A.; Li, W.; Anderson, E.L.; Wong, E.H.M.; Dulai, P.S.; Sandborn, W.J.; Biggs, W.; Yooseph, S.; Jones, M.B.; Venter, C.J.; et al. Genetic risk, dysbiosis, and treatment stratification using host genome and gut microbiome in inflammatory bowel disease. Clin. Transl. Gastroenterol. 2018, 9, e132. [Google Scholar] [CrossRef]
- Cao, Y.; Shen, J.; Ran, Z.H. Association between Faecalibacterium prausnitzii Reduction and Inflammatory Bowel Disease: A Meta-Analysis and Systematic Review of the Literature. Gastroenterol. Res. Pr. 2014, 2014, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Rossi, O.; Van Berkel, L.A.; Chain, F.; Khan, M.T.; Taverne, N.; Sokol, H.; Duncan, S.H.; Flint, H.J.; Harmsen, H.J.M.; Langella, P.; et al. Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci. Rep. 2016, 6, 18507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srutkova, D.; Schwarzer, M.; Hudcovic, T.; Zakostelska, Z.; Drab, V.; Spanova, A.; Rittich, B.; Kozakova, H.; Schabussova, I. Bifidobacterium longum CCM 7952 Promotes Epithelial Barrier Function and Prevents Acute DSS-Induced Colitis in Strictly Strain-Specific Manner. PLoS ONE 2015, 10, e0134050. [Google Scholar] [CrossRef]
- Fehlbaum, S.; Prudence, K.; Kieboom, J.; Heerikhuisen, M.; Broek, T.V.D.; Schuren, F.H.J.; Steinert, R.E.; Raederstorff, D. In Vitro Fermentation of Selected Prebiotics and Their Effects on the Composition and Activity of the Adult Gut Microbiota. Int. J. Mol. Sci. 2018, 19, 3097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinsbroek, S.E.; Oei, A.; Roelofs, J.J.T.H.; Dhawan, S.; Velde, A.T.; Gordon, S.; De Jonge, W.J. Genetic deletion of dectin-1 does not affect the course of murine experimental colitis. BMC Gastroenterol. 2012, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Hove, T.T.; Drillenburg, P.; Wijnholds, J.; Velde, A.A.T.; Van Deventer, S.J. Differential susceptibility of multidrug resistance protein-1 deficient mice to DSS and TNBS-induced colitis. Dig. Dis. Sci. 2002, 47, 2056–2063. [Google Scholar] [CrossRef] [PubMed]
- Cooper, H.S.; Murthy, S.N.; Shah, R.S.; Sedergran, D.J. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Investig. 1993, 69, 238–249. [Google Scholar] [PubMed]
- Heinsbroek, S.E.; Williams, D.L.; Welting, O.; Meijer, S.L.; Gordon, S.; De Jonge, W.J. Orally delivered β-glucans aggravate dextran sulfate sodium (DSS)-induced intestinal inflammation. Nutr. Res. 2015, 35, 1106–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willemze, R.A.; Brinkman, D.J.; Welting, O.; Van Hamersveld, P.H.P.; Verseijden, C.; Luyer, M.D.; Wildenberg, M.E.; Seppen, J.; De Jonge, W.J.; Van Hamersveld, H.P. Acetylcholine-producing T cells augment innate immune-driven colitis but are redundant in T cell-driven colitis. Am. J. Physiol. Liver Physiol. 2019, 317, G557–G568. [Google Scholar] [CrossRef] [PubMed]
- Willemze, R.A.; Welting, O.; Van Hamersveld, H.P.; Meijer, S.L.; Folgering, A.J.H.; Darwinkel, H.; Witherington, J.; Sridhar, A.; Vervoordeldonk, M.J.; Seppen, J.; et al. Neuronal control of experimental colitis occurs via sympathetic intestinal innervation. Neurogastroenterol. Motil. 2017, 30, e13163. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; Hoff, M.J.B.V.D.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladirat, S.E.; Schols, H.A.; Nauta, A.; Schoterman, M.H.C.; Keijser, B.J.F.; Montijn, R.C.; Gruppen, H.; Schuren, F.H.J. High-throughput analysis of the impact of antibiotics on the human intestinal microbiota composition. J. Microbiol. Methods 2013, 92, 387–397. [Google Scholar] [CrossRef]
- Haak, B.W.; Argelaguet, R.; Kinsella, C.M.; Kullberg, R.F.J.; Lankelma, J.M.; Deijs, M.; Klein, M.; Jebbink, M.F.; Hugenholtz, F.; Kostidis, S.; et al. Integrative Transkingdom Analysis of the Gut Microbiome in Antibiotic Perturbation and Critical Illness. mSystems 2021, 6. [Google Scholar] [CrossRef]
- Edgar, R. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 2016, 081257. [Google Scholar] [CrossRef] [Green Version]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 1–14. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Schliep, K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package; GitHub: San Francisco, CA, USA, 2019. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okayasu, I.; Hatakeyama, S.; Yamada, M.; Ohkusa, T.; Inagaki, Y.; Nakaya, R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology 1990, 98, 694–702. [Google Scholar] [CrossRef]
- Rice, P.J.; Adams, E.L.; Ozment-Skelton, T.; Gonzalez, A.J.; Goldman, M.P.; Lockhart, B.E.; Barker, L.A.; Breuel, K.F.; Deponti, W.K.; Kalbfleisch, J.H.; et al. Oral Delivery and Gastrointestinal Absorption of Soluble Glucans Stimulate Increased Resistance to Infectious Challenge. J. Pharmacol. Exp. Ther. 2005, 314, 1079–1086. [Google Scholar] [CrossRef]
- Larmonier, C.B.; Laubitz, D.; Hill, F.M.; Shehab, K.W.; Lipinski, L.; Midura-Kiela, M.T.; McFadden, R.-M.T.; Ramalingam, R.; Hassan, K.A.; Golebiewski, M.; et al. Reduced colonic microbial diversity is associated with colitis in NHE3-deficient mice. Am. J. Physiol. Liver Physiol. 2013, 305, G667–G677. [Google Scholar] [CrossRef] [Green Version]
- De Fazio, L. Longitudinal analysis of inflammation and microbiota dynamics in a model of mild chronic dextran sulfate sodium-induced colitis in mice. World J. Gastroenterol. 2014, 20, 2051–2061. [Google Scholar] [CrossRef]
- Munyaka, P.M.; Rabbi, M.F.; Khafipour, E.; Ghia, J.-E. Acute dextran sulfate sodium (DSS)-induced colitis promotes gut microbial dysbiosis in mice. J. Basic Microbiol. 2016, 56, 986–998. [Google Scholar] [CrossRef]
- Gao, Z.; Chen, K.-Y.; Mueller, O.; Zhang, H.; Rakhilin, N.; Chen, J.; Shen, X. Microbiota of Inflammatory Bowel Disease Models. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; Volume 2018, pp. 2374–2377. [Google Scholar]
- Pei, L.-Y.; Ke, Y.-S.; Zhao, H.-H.; Wang, L.; Jia, C.; Liu, W.-Z.; Fu, Q.-H.; Shi, M.-N.; Cui, J.; Li, S.-C. Role of colonic microbiota in the pathogenesis of ulcerative colitis. BMC Gastroenterol. 2019, 19, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Duranti, S.; Gaiani, F.; Mancabelli, L.; Milani, C.; Grandi, A.; Bolchi, A.; Santoni, A.; Lugli, G.A.; Ferrario, C.; Mangifesta, M.; et al. Elucidating the gut microbiome of ulcerative colitis: Bifidobacteria as novel microbial biomarkers. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Suolang, Y.; Zhou, D.; Tang, Y.; Zhang, Y. Bifidobacteria alleviate experimentally induced colitis by upregulating indoleamine 2, 3-dioxygenase expression. Microbiol. Immunol. 2018, 62, 71–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, B. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis: A role for bifidobacteria and lactobacilli? Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 205. [Google Scholar] [CrossRef] [Green Version]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, D.-H.; Seo, D.-H.; Kim, G.-Y.; Nam, Y.-D.; Song, E.-J.; Yoon, S.; Park, C.-S. The effect of resistant starch (RS) on the bovine rumen microflora and isolation of RS-degrading bacteria. Appl. Microbiol. Biotechnol. 2018, 102, 4927–4936. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, J.; Schlormann, W.; Trautvetter, U.; Glei, M. The effects of β-glucans on intestinal health. Ernahrungs Umschau 2020, 67, 52–59. [Google Scholar]
- Shi, L.; Lin, Q.; Yang, T.; Nie, Y.; Li, X.; Liu, B.; Shen, J.; Liang, Y.; Tang, Y.; Luo, F. Oral administration of Lentinus edodes β-glucans ameliorates DSS-induced ulcerative colitis in mice via MAPK-Elk-1 and MAPK-PPARγ pathways. Food Funct. 2016, 7, 4614–4627. [Google Scholar] [CrossRef]
- Liu, B.; Lin, Q.; Yang, T.; Zeng, L.; Shi, L.; Chen, Y.; Luo, F. Oat β-glucan ameliorates dextran sulfate sodium (DSS)-induced ulcerative colitis in mice. Food Funct. 2015, 6, 3454–3463. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, Z.; Chen, J.; Zhan, Y.; Wang, T.; Xia, L.; Wang, S.; Hua, Z.; Zhang, J. Supplementation of the diet with Salecan attenuates the symptoms of colitis induced by dextran sulphate sodium in mice. Br. J. Nutr. 2014, 111, 1822–1829. [Google Scholar] [CrossRef] [Green Version]
- Petersson, J.; Schreiber, O.; Hansson, G.C.; Gendler, S.J.; Velcich, A.; Lundberg, J.O.; Roos, S.; Holm, L.; Phillipson, M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. Liver Physiol. 2011, 300, G327–G333. [Google Scholar] [CrossRef] [Green Version]
- Kiesler, P.; Fuss, I.J.; Strober, W. Experimental Models of Inflammatory Bowel Diseases. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 154–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Meer, J.W.M.; Joosten, L.A.B.; Riksen, N.; Netea, M.G. Trained immunity: A smart way to enhance innate immune defence. Mol. Immunol. 2015, 68, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Quintin, J. Fungal mediated innate immune memory, what have we learned? Semin. Cell Dev. Biol. 2019, 89, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Camilli, G.; Bohm, M.; Piffer, A.C.; Lavenir, R.; Williams, D.L.; Neven, B.; Grateau, G.; Georgin-Lavialle, S.; Quintin, J. β-Glucan–induced reprogramming of human macrophages inhibits NLRP3 inflammasome activation in cryopyrinopathies. J. Clin. Investig. 2020, 130, 4561–4573. [Google Scholar] [CrossRef]
- Paris, S.; Chapat, L.; Pasin, M.; Lambiel, M.; Sharrock, T.E.; Shukla, R.; Sigoillot-Claude, C.; Bonnet, J.-M.; Poulet, H.; Freyburger, L.; et al. β-Glucan-Induced Trained Immunity in Dogs. Front. Immunol. 2020, 11, 566893. [Google Scholar] [CrossRef]
- Lührs, H.; Gerke, T.; Muller, J.G.; Melcher, R.; Schauber, J.; Boxberge, F.; Scheppach, W.; Menzel, T. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand. J. Gastroenterol. 2002, 37, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Lee, E.-J.; Lee, J.-C.; Kim, W.-K.; Kim, H.-S. Anti-inflammatory effects of short chain fatty acids in IFN-gamma-stimulated RAW 264.7 murine macrophage cells: Involvement of NF-kappaB and ERK signaling pathways. Int. Immunopharmacol. 2007, 7, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayachandran, M.; Chen, J.; Chung, S.S.M.; Xu, B. A critical review on the impacts of β-glucans on gut microbiota and human health. J. Nutr. Biochem. 2018, 61, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Gudi, R.; Suber, J.; Brown, R.; Johnson, B.M.; Vasu, C. Pretreatment with Yeast-Derived Complex Dietary Polysaccharides Suppresses Gut Inflammation, Alters the Microbiota Composition, and Increases Immune Regulatory Short-Chain Fatty Acid Production in C57BL/6 Mice. J. Nutr. 2019, 150, 1291–1302. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Li, J.; Liu, Y.; Yue, W.; Luo, X. Toll-like receptor 2 monoclonal antibody or/and Toll-like receptor 4 monoclonal antibody increase counts of Lactobacilli and Bifidobacteria in dextran sulfate sodium-induced colitis in mice. J. Gastroenterol. Hepatol. 2011, 27, 110–119. [Google Scholar] [CrossRef]
- Chae, J.M.; Heo, W.; Cho, H.T.; Lee, D.H.; Kim, J.H.; Rhee, M.S.; Park, T.-S.; Kim, Y.K.; Lee, J.H.; Kim, Y.J. Effects of Orally-Administered Bifidobacterium animalis subsp. lactis Strain BB12 on Dextran Sodium Sulfate-Induced Colitis in Mice. J. Microbiol. Biotechnol. 2018, 28, 1800–1805. [Google Scholar] [CrossRef]
- Macfarlane, S.; Furrie, E.; Cummings, J.H.; Macfarlane, G.T. Chemotaxonomic Analysis of Bacterial Populations Colonizing the Rectal Mucosa in Patients with Ulcerative Colitis. Clin. Infect. Dis. 2004, 38, 1690–1699. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, F.J.; Duncan, S.H.; Hold, G.; Stewart, C.S. Isolation, growth on prebiotics and probiotic potential of novel bifidobacteria from pigs. Anaerobe 2004, 10, 33–39. [Google Scholar] [CrossRef]
- Jung, D.-H.; Chung, W.-H.; Seo, D.-H.; Nam, Y.-D.; Yoon, S.; Park, C.-S. Complete genome sequence of Bifidobacterium choerinum FMB-1, a resistant starch-degrading bacterium. J. Biotechnol. 2018, 274, 28–32. [Google Scholar] [CrossRef]
- Ott, S.J.; Kühbacher, T.; Musfeldt, M.; Rosenstiel, P.; Hellmig, S.; Rehman, A.; Drews, O.; Weichert, W.; Timmis, K.N.; Schreiber, S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand. J. Gastroenterol. 2008, 43, 831–841. [Google Scholar] [CrossRef]
- Underhill, D.M.; Iliev, I.D. The mycobiota: Interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 2014, 14, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions Between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.; Zhang, F.; Yang, X.; Wu, N.; Jiang, W.; Li, X.; Li, X.; Liu, Y. Changes in the composition of intestinal fungi and their role in mice with dextran sulfate sodium-induced colitis. Sci. Rep. 2015, 5, 10416. [Google Scholar] [CrossRef]
- Frank, D.N.; Amand, A.L.S.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [Green Version]
- Loh, G.; Blaut, M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes 2012, 3, 544–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Zhi, F. Lower Level of Bacteroides in the Gut Microbiota Is Associated with Inflammatory Bowel Disease: A Meta-Analysis. BioMed Res. Int. 2016, 2016, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Devaux, C.A.; Million, M.; Raoult, D. The Butyrogenic and Lactic Bacteria of the Gut Microbiota Determine the Outcome of Allogenic Hematopoietic Cell Transplant. Front. Microbiol. 2020, 11, 1642. [Google Scholar] [CrossRef]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R.; et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 1–12. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2016, 66, 1039–1048. [Google Scholar] [CrossRef] [Green Version]
- Jenq, R.R.; Taur, Y.; Devlin, S.M.; Ponce, D.M.; Goldberg, J.D.; Ahr, K.F.; Littmann, E.R.; Ling, L.; Gobourne, A.C.; Miller, L.C.; et al. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2015, 21, 1373–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paramsothy, S.; Kamm, A.M.; Kaakoush, O.N.; Walsh, A.J.; Bogaerde, J.V.D.; Samuel, D.; Leong, R.W.L.; Connor, S.; Ng, W.; Paramsothy, R.; et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: A randomised placebo-controlled trial. Lancet 2017, 389, 1218–1228. [Google Scholar] [CrossRef]






| Gene | 5′-Forward Sequence | 5′-Reverse Sequence |
|---|---|---|
| GAPDH | ATGTGTCCGTCGTGGATCTGA | ATGCCTGCTTCACCACCTTCT |
| Cyclophilin | ATGGTCAACCCCACCGTGT | TTCTGCTGTCTTTGGAACTTTGTC |
| RPLP0 | CCAGCGAGGCCACACTGCTG | ACACTGGCCACGTTGCGGAC |
| TNF-α | AAAGCATGATCCGCGACGT | TGCAAGCAGGAATGAGAA |
| IL-6 | GAGTTGTGCAATGGCAATTCTG | TGGTAGCATCCATCATTTCTTTGT |
| IL-10 | TGTCAAATTCATTCATGGCCT | ATCGATTTCTCCCCTGTGAA |
| NOS2 | TTCTGTGCTGTCCCAGTGAG | TGAAGAAAACCCCTTGTGCT |
| CCL-2 | AGGTCCCTGTCATGCTTCTG | TCTCCAGCCTACTCATTGGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, S.; Davids, M.; van Hamersveld, P.H.P.; Welting, O.; Rahaoui, H.; Schuren, F.; Meijer, S.L.; van den Wijngaard, R.M.; Hakvoort, T.B.M.; de Jonge, W.J.; et al. Dietary Curdlan Enhances Bifidobacteria and Reduces Intestinal Inflammation in Mice. Nutrients 2021, 13, 1305. https://doi.org/10.3390/nu13041305
Rahman S, Davids M, van Hamersveld PHP, Welting O, Rahaoui H, Schuren F, Meijer SL, van den Wijngaard RM, Hakvoort TBM, de Jonge WJ, et al. Dietary Curdlan Enhances Bifidobacteria and Reduces Intestinal Inflammation in Mice. Nutrients. 2021; 13(4):1305. https://doi.org/10.3390/nu13041305
Chicago/Turabian StyleRahman, Shafaque, Mark Davids, Patricia H. P. van Hamersveld, Olaf Welting, Hakim Rahaoui, Frank Schuren, Sybren L. Meijer, René M. van den Wijngaard, Theodorus B. M. Hakvoort, Wouter J. de Jonge, and et al. 2021. "Dietary Curdlan Enhances Bifidobacteria and Reduces Intestinal Inflammation in Mice" Nutrients 13, no. 4: 1305. https://doi.org/10.3390/nu13041305