From Space to Field: Combining Satellite, UAV and Agronomic Data in an Open-Source Methodology for the Validation of NDVI Maps in Precision Viticulture
Abstract
:1. Introduction
2. Materials and Methods
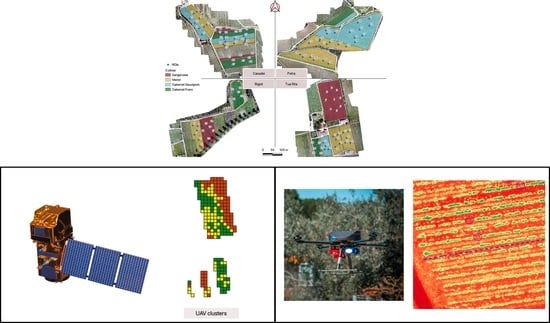
2.1. Monitoring Sites, Plant Material and Weather Data
2.2. Satellite Images
2.3. Unmanned Aerial Vehicles Images
2.4. Images Alignment, Segmentation and Consistency Analysis
- NDVI S2: NDVI maps derived from Sentinel-2 multi-spectral images;
- NDVI UAVplain: NDVI maps derived from full UAV multi-spectral images;
- NDVI UAVvines: NDVI maps derived segmenting vines-only pixels from UAVplain images.
2.5. Comparative Spatial Analyses
2.6. In-Field Validation and Ground Data
3. Results
3.1. Classical Statistical Approach: Ordinary Least Square Regression
3.2. Spatial Statistical Analysis: Bivariate Moran’s Index
3.3. Distribution Assessment of Agronomic Variables over NDVI Clusters
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of Variance |
| GIS | Geographic Information System |
| GCPs | Ground Control Points |
| LWA | Leaf Wall Area |
| MI | Moran’s Index |
| NDVI | Normalized Difference Vegetation Index |
| OLS | Ordinary Least Squares regression |
| ROI | Region of Interest |
| PA | Precision Agriculture |
| PV | Precision Viticulture |
| SC | Scene Classification |
| UAV | Unmanned Aerial Vehicles |
Appendix A. NDVI Clusters






References
- Crookston, R.K. A top 10 list of developments and issues impacting crop management and ecology during the past 50 years. Crop Sci. 2006, 46, 2253–2262. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Santesteban, L.G. Precision viticulture and advanced analytics. A short review. Food Chem. 2019, 279, 58–62. [Google Scholar] [CrossRef]
- ISTAT. Stima Sulla Superficie Vitivinicola e Produzione Vinicola; ISTAT: Roma, Italy, 2021.
- Malone, P.; Apgar, H.; Stukes, S.; Sterk, S. Unmanned aerial vehicles unique cost estimating requirements. In Proceedings of the 2013 IEEE Aerospace Conference, Big Sky, MT, USA, 2–9 March 2013; pp. 1–8. [Google Scholar]
- D’Odorico, P.; Gonsamo, A.; Damm, A.; Schaepman, M.E. Experimental evaluation of Sentinel-2 spectral response functions for NDVI time-series continuity. IEEE Trans. Geosci. Remote Sens. 2013, 51, 1336–1348. [Google Scholar] [CrossRef]
- Segarra, J.; Buchaillot, M.L.; Araus, J.L.; Kefauver, S.C. Remote sensing for precision agriculture: Sentinel-2 improved features and applications. Agronomy 2020, 10, 641. [Google Scholar] [CrossRef]
- Rouse, J.W., Jr.; Haas, R.H.; Deering, D.; Schell, J.; Harlan, J.C. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation. No. E75-10354. 1974. Available online: https://ntrs.nasa.gov/citations/19730017588 (accessed on 2 May 2023).
- Huang, S.; Tang, L.; Hupy, J.P.; Wang, Y.; Shao, G. A commentary review on the use of normalized difference vegetation index (NDVI) in the era of popular remote sensing. J. For. Res. 2021, 32, 1–6. [Google Scholar] [CrossRef]
- Sozzi, M.; Kayad, A.; Marinello, F.; Taylor, J.; Tisseyre, B. Comparing vineyard imagery acquired from Sentinel-2 and Unmanned Aerial Vehicle (UAV) platform. Oeno One 2020, 54, 189–197. [Google Scholar] [CrossRef]
- Nonni, F.; Malacarne, D.; Pappalardo, S.E.; Codato, D.; Meggio, F.; De Marchi, M. Sentinel-2 Data Analysis and Comparison with UAV Multispectral Images for Precision Viticulture. GI Forum 2018, 1, 105–116. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Dainelli, R.; Palliotti, A.; Toscano, P.; Matese, A. Sentinel-2 validation for spatial variability assessment in overhead trellis system viticulture versus UAV and agronomic data. Remote Sens. 2019, 11, 2573. [Google Scholar] [CrossRef]
- Khaliq, A.; Comba, L.; Biglia, A.; Ricauda Aimonino, D.; Chiaberge, M.; Gay, P. Comparison of satellite and UAV-based multispectral imagery for vineyard variability assessment. Remote Sens. 2019, 11, 436. [Google Scholar] [CrossRef]
- Pastonchi, L.; Di Gennaro, S.F.; Toscano, P.; Matese, A. Comparison between satellite and ground data with UAV-based information to analyse vineyard spatio-temporal variability: This article is published in cooperation with the XIIIth International Terroir Congress November 17-18 2020, Adelaide, Australia. Guest editors: Cassandra Collins and Roberta De Bei. Oeno One 2020, 54, 919–934. [Google Scholar]
- Matese, A.; Di Gennaro, S.F.; Santesteban, L.G. Methods to compare the spatial variability of UAV-based spectral and geometric information with ground autocorrelated data. A case of study for precision viticulture. Comput. Electron. Agric. 2019, 162, 931–940. [Google Scholar] [CrossRef]
- Walklate, P.; Cross, J. An examination of Leaf-Wall-Area dose expression. Crop Prot. 2012, 35, 132–134. [Google Scholar] [CrossRef]
- Spectator. Available online: http://app.spectator.earth (accessed on 2 May 2023).
- CNR Institute of Atmospheric Sciences and Climate—Climate Monitoring for Italy. Available online: https://www.isac.cnr.it/climstor/ (accessed on 2 May 2023).
- Weather Underground. Available online: https://www.wunderground.com/ (accessed on 2 May 2023).
- Copernicus Open Access Hub. Available online: https://scihub.copernicus.eu/ (accessed on 2 May 2023).
- QGIS Development Team. QGIS Geographic Information System; Open Source Geospatial Foundation: Chicago, IL, USA, 2009. [Google Scholar]
- Xavier Corredor Llano. Coregistration QGIS Plugin; Colombian Forest Monitoring System, SMByC: Colombia, 2021; Available online: https://github.com/SMByC/ (accessed on 2 May 2023).
- Devaux, N.; Crestey, T.; Leroux, C.; Tisseyre, B. Potential of Sentinel-2 satellite images to monitor vine fields grown at a territorial scale. Oeno One 2019, 53, 52–59. [Google Scholar] [CrossRef]
- Barros, T.; Conde, P.; Gonçalves, G.; Premebida, C.; Monteiro, M.; Ferreira, C.S.S.; Nunes, U.J. Multispectral vineyard segmentation: A deep learning comparison study. Comput. Electron. Agric. 2022, 195, 106782. [Google Scholar] [CrossRef]
- Cinat, P.; Di Gennaro, S.F.; Berton, A.; Matese, A. Comparison of unsupervised algorithms for Vineyard Canopy segmentation from UAV multispectral images. Remote Sens. 2019, 11, 1023. [Google Scholar] [CrossRef]
- Moran, P.A. Notes on continuous stochastic phenomena. Biometrika 1950, 37, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Anselin, L.; Syabri, I.; Kho, Y. GeoDa: An introduction to spatial data analysis. In Handbook of Applied Spatial Analysis: Software Tools, Methods and Applications; Springer: Berlin/Heidelberg, Germany, 2009; pp. 73–89. [Google Scholar]
- Taylor, J.; Bates, T. A discussion on the significance associated with Pearson’s correlation in precision agriculture studies. Precis. Agric. 2013, 14, 558–564. [Google Scholar] [CrossRef]
- Anselin, L.; Rey, S.J. Modern Spatial Econometrics in Practice: A Guide to GeoDa, GeoDaSpace and PySAL. 2014. Available online: https://sergerey.org/giasp16/pdfs/anselin_rey_weights.pdf (accessed on 2 May 2023).
- St, L.; Wold, S. Analysis of variance (ANOVA). Chemom. Intell. Lab. Syst. 1989, 6, 259–272. [Google Scholar]
- Ugoni, A.; Walker, B.F. The Chi square test: An introduction. COMSIG Rev. 1995, 4, 61. [Google Scholar]









| Winery | Soil Texture | Soil Management | Layout (m) | Cultivar | Extension (ha) | Training Method |
|---|---|---|---|---|---|---|
| Casadei | Silty clay loam | Alternated cover crop | 1.80 × 0.80 | Sangiovese | 0.45 | Double spurred cordon |
| Merlot | 1.21 | Spurred Cordon | ||||
| Cabernet Sauvignon | 1.11 | Spurred Cordon | ||||
| Cabernet Franc | 1.24 | Double Spurred Cordon | ||||
| Petra | Clay loam | Alternated cover crop | 1.60 × 0.80 | Sangiovese | / | / |
| Merlot | 2.82 | Guyot | ||||
| Cabernet Sauvignon | 3.39 | Guyot | ||||
| Cabernet Franc | 0.47 | Spurred Cordon | ||||
| Rigoli | Sandy clay loam | Spontaneous grassing | 2.20 × 0.80 | Sangiovese | 0.88 | Spurred Cordon |
| Merlot | 0.76 | Spurred Cordon | ||||
| Cabernet Sauvignon | / | / | ||||
| Cabernet Franc | 0.62 | Guyot | ||||
| Tua Rita | Loam | Alternated cover crop | 1.40 × 0.80 | Sangiovese | 2.22 | Double Spurred Cordon |
| Merlot | 1.49 | Double Spurred Cordon | ||||
| Cabernet Sauvignon | 0.41 | Double Spurred Cordon | ||||
| Cabernet Franc | 0.27 | Double Spurred Cordon |
| Flight | Date |
|---|---|
| F1 | 24 March 2022 |
| F2 | 28 April 2022 |
| F3 | 23 May 2022 |
| F4 | 27 June 2022 |
| F5 | 22 July 2022 |
| F6 | 26 August 2022 |
| F1 | F2 | F3 | F4 | F5 | F6 | |
|---|---|---|---|---|---|---|
| Casadei | On alternated rows | Just mowed | Absent | Absent | Absent | Absent |
| Petra | On alternated rows, uneven | On alternated rows, uneven | Absent | Absent | Absent | Partially present |
| Rigoli | Spontaneous | Just mowed | Just mowed | Absent | Absent | Partially present |
| Tua Rita | On alternated rows | Just mowed | Absent | Absent | Absent | Absent |
| Focal distance | 5.5 mm |
| GSD | 8 cm/pixel (per band) at 120 m AGL |
| Field of View | 47.2° HFOV |
| Resolution | 1280 × 960 |
| B (475 nm ± 20 nm) | |
| G (560 nm ± 20 nm) | |
| Spectral bands | R (668 nm ± 10 nm) |
| Red Edge (717 nm ± 10 nm) | |
| NIR (840 nm ± 40 nm) |
| Site | F1 | F2 | F3 | F4 | F5 | F6 | |
|---|---|---|---|---|---|---|---|
| Univariate MI for NDVI S2 | Casadei | 0.848 | / | 0.769 | 0.802 | 0.857 | 0.857 |
| Petra | 0.901 | 0.868 | / | 0.719 | 0.758 | 0.785 | |
| Rigoli | 0.662 | 0.851 | 0.934 | 0.882 | 0.887 | 0.867 | |
| Tua Rita | / | 0.743 | 0.753 | 0.673 | 0.855 | 0.863 | |
| Univariate MI for NDVI UAVplain | Casadei | 0.624 | / | 0.718 | 0.859 | 0.774 | 0.756 |
| Petra | 0.845 | 0.812 | / | 0.640 | 0.706 | 0.699 | |
| Rigoli | 0.626 | 0.700 | 0.856 | 0.759 | 0.852 | 0.825 | |
| Tua Rita | / | 0.673 | 0.785 | 0.718 | 0.736 | 0.433 | |
| Univariate MI for NDVI UAVvines | Casadei | 0.695 | / | 0.566 | 0.831 | 0.777 | 0.856 |
| Petra | 0.817 | 0.833 | / | 0.684 | 0.717 | 0.778 | |
| Rigoli | 0.793 | 0.898 | 0.883 | 0.545 | 0.960 | 0.675 | |
| Tua Rita | / | 0.682 | 0.637 | 0.823 | 0.726 | 0.717 |
| F1 | F2 | F3 | F4 | F5 | F6 | |
|---|---|---|---|---|---|---|
| Casadei | 0.709 | / | 0.808 | 0.745 | 0.778 | 0.814 |
| Petra | 0.920 | 0.902 | / | 0.703 | 0.721 | 0.792 |
| Rigoli | 0.802 | 0.668 | 0.890 | 0.693 | 0.825 | 0.642 |
| Tua Rita | / | 0.338 | 0.589 | 0.693 | 0.825 | 0.642 |
| F1 | F2 | F3 | F4 | F5 | F6 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Casadei | / | / | / | / | 0.435 | 0.660 | 0.655 | 0.809 | 0.715 | 0.845 | 0.801 | 0.895 |
| Petra | / | / | 0.527 | 0.726 | / | / | 0.118 | 0.343 | 0.041 | 0.203 | 0.287 | 0.536 |
| Rigoli | / | / | 0.625 | 0.790 | 0.787 | 0.887 | 0.256 | 0.506 | 0.469 | 0.684 | 0.678 | 0.823 |
| TuaRita | / | / | 0.369 | 0.607 | 0.152 | 0.389 | 0.396 | 0.629 | 0.674 | 0.821 | 0.763 | 0.873 |
| F1 | F2 | F3 | F4 | F5 | F6 | |
|---|---|---|---|---|---|---|
| Casadei | 0.733 | / | 0.719 | 0.773 | 0.797 | 0.799 |
| Petra | 0.853 | 0.806 | / | 0.622 | 0.640 | 0.701 |
| Rigoli | 0.638 | 0.750 | 0.869 | 0.771 | 0.822 | 0.714 |
| Tua Rita | / | 0.348 | 0.679 | 0.616 | 0.776 | 0.439 |
| F2 | F3 | F4 | F5 | F6 | |
|---|---|---|---|---|---|
| Casadei | / | 0.546 | 0.737 | 0.765 | 0.836 |
| Petra | 0.628 | / | 0.189 | 0.057 | 0.417 |
| Rigoli | 0.757 | 0.840 | 0.430 | 0.639 | 0.747 |
| Tua Rita | 0.411 | 0.342 | 0.509 | 0.691 | 0.765 |
| Casadei | Petra | Rigoli | Tua Rita | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | Medium | High | Low | Medium | High | Low | Medium | High | Low | Medium | High | |
| F2 | 9.83 | 10.5 | 14.83 | 8.8 | 13.18 | 14.27 | 5.13 | 3.67 | 7.14 | 8 | 9.9 | 12.67 |
| F3 | 47.86 | 59.57 | 63 | 74 | 80 | 86 | 42.5 | 53.29 | 62 | 54.83 | 53.5 | 61.25 |
| F4 | 64.75 | 70.40 | 90.33 | 90.38 | 98.92 | 105 | 75 | 92.43 | 93.33 | 60.29 | 62.86 | 70.60 |
| F5 | 69.86 | 68.40 | 71.75 | 97.50 | 98.17 | 98.56 | 77.50 | 93.86 | 96.67 | 57.89 | 62.17 | 65.67 |
| F6 | 63.83 | 71.86 | 92 | 100.60 | 93.36 | 96 | 91 | 95.75 | 93.33 | 60.38 | 62 | 62.62 |
| Casadei | Petra | Rigoli | Tua Rita | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | Medium | High | Low | Medium | High | Low | Medium | High | Low | Medium | High | |
| F2 | 10.43 | 10.75 | 14.8 | 9.17 | 11.83 | 14.67 | 4.05 | 5.7 | 7.8 | 7 | 8.78 | 13.11 |
| F3 | 47.86 | 57.83 | 64.40 | 74.38 | 84.29 | 84.09 | 42.4 | 50.57 | 62 | 52 | 54.75 | 64 |
| F4 | 59.14 | 76.71 | 95 | 93.17 | 91.7 | 106.27 | 75.43 | 97.80 | 90 | 58.60 | 64.43 | 67.29 |
| F5 | 62.4 | 72 | 75.5 | 108 | 90.89 | 97.82 | 79.29 | 94.4 | 96.25 | 58.11 | 63.67 | 64.44 |
| F6 | 59.20 | 75.86 | 83.75 | 95.57 | 94.36 | 96.78 | 89.5 | 97.25 | 93.33 | 57.5 | 62.88 | 67 |
| Casadei | Petra | Rigoli | Tua Rita | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | Medium | High | Low | Medium | High | Low | Medium | High | Low | Medium | High | |
| F2 | 8.5 | 11.17 | 14.83 | 8.88 | 9.33 | 16.85 | 3 | 5 | 9.36 | 4.83 | 9.14 | 13.27 |
| F3 | 47.86 | 57.71 | 66.25 | 64.5 | 80.23 | 97.14 | 42 | 51.75 | 62.67 | 45.5 | 56.25 | 64.63 |
| F4 | 61.57 | 64.80 | 96.5 | 83.86 | 96.10 | 109.70 | 73.3 | 89.8 | 97.6 | 58.8 | 64.33 | 70 |
| F5 | 56.14 | 79.83 | 82 | 94.67 | 97.22 | 102.56 | 74 | 94 | 95.60 | 56.67 | 64.14 | 65.75 |
| F6 | 56.80 | 78.8 | 90 | 92.75 | 95.5 | 97.89 | 92.8 | 94.33 | 93.33 | 58.57 | 62.20 | 63.25 |
| Casadei | Petra | Rigoli | Tua Rita | |||||
|---|---|---|---|---|---|---|---|---|
| F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | |
| F2 | 3.359 | 0.067 | 2.202 | 0.132 | 1.802 | 0.204 | 1.758 | 0.197 |
| F3 | 3.854 | 0.045 | 0.637 | 0.538 | 5.982 | 0.014 | 0.455 | 0.640 |
| F4 | 1.504 | 0.259 | 2.185 | 0.134 | 3.476 | 0.062 | 1.836 | 0.192 |
| F5 | 0.023 | 0.978 | 0.008 | 0.992 | 4.707 | 0.029 | 2.664 | 0.093 |
| F6 | 2.405 | 0.129 | 0.729 | 0.493 | 0.249 | 0.786 | 0.185 | 0.833 |
| Casadei | Petra | Rigoli | Tua Rita | |||||
|---|---|---|---|---|---|---|---|---|
| F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | |
| F2 | 2.169 | 0.154 | 3.029 | 0.067 | 2.634 | 0.11 | 4.427 | 0.025 |
| F3 | 4.582 | 0.028 | 0.671 | 0.521 | 6.293 | 0.012 | 1.836 | 0.184 |
| F4 | 2.915 | 0.09 | 3.849 | 0.035 | 5.465 | 0.019 | 1.179 | 0.333 |
| F5 | 0.42 | 0.665 | 2.963 | 0.071 | 3.914 | 0.047 | 1.895 | 0.175 |
| F6 | 2.155 | 0.156 | 0.102 | 0.903 | 0.739 | 0.508 | 4.193 | 0.029 |
| Casadei | Petra | Rigoli | Tua Rita | |||||
|---|---|---|---|---|---|---|---|---|
| F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | |
| F2 | 4.42 | 0.034 | 18.014 | <0.001 | 25.897 | <0.001 | 12.496 | <0.001 |
| F3 | 5.212 | 0.019 | 6.459 | 0.006 | 5.323 | 0.02 | 5.918 | 0.009 |
| F4 | 5.259 | 0.021 | 13.433 | <0.001 | 6.649 | 0.01 | 7.002 | 0.007 |
| F5 | 3.159 | 0.076 | 0.632 | 0.54 | 8.056 | 0.005 | 4.432 | 0.025 |
| F6 | 3.172 | 0.076 | 0.405 | 0.672 | 0.023 | 0.977 | 0.901 | 0.421 |
| Casadei | Petra | Rigoli | Tua Rita | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | Medium | High | Low | Medium | High | Low | Medium | High | Low | Medium | High | |
| F3 | 32.15 | 47.76 | 45.63 | 72.96 | 87.83 | 93.43 | 23.93 | 48.57 | 54.27 | 49.21 | 42.65 | 50.87 |
| F4 | 59.94 | 59.03 | 98.66 | 92.67 | 106.72 | 107.16 | 58.27 | 87.74 | 90.18 | 49.05 | 59.22 | 70.31 |
| F5 | 69.36 | 66.82 | 67.12 | 95.24 | 100.87 | 101.14 | 54.98 | 85.03 | 93.53 | 50.57 | 57.39 | 66.94 |
| F6 | 63.32 | 72.23 | 79.39 | 96 | 96.19 | 96.06 | 85.58 | 98.17 | 96.28 | 50.19 | 61.73 | 66.38 |
| Casadei | Petra | Rigoli | Tua Rita | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | Medium | High | Low | Medium | High | Low | Medium | High | Low | Medium | High | |
| F3 | 32.15 | 46.51 | 46.51 | 74.84 | 98.18 | 89.02 | 23.56 | 43.54 | 55.96 | 46.14 | 39.94 | 54.92 |
| F4 | 46.64 | 78.38 | 97.78 | 97.75 | 95.28 | 112.07 | 59.54 | 101.49 | 77.51 | 51.54 | 55.32 | 66.37 |
| F5 | 66.85 | 69.01 | 67.68 | 103.13 | 97.34 | 99.47 | 59.99 | 83.94 | 91.52 | 48.18 | 59.85 | 67.69 |
| F6 | 62.69 | 70.15 | 79.79 | 101.28 | 94.7 | 93.83 | 85.48 | 89.08 | 108.53 | 47.22 | 63.91 | 73.8 |
| Casadei | Petra | Rigoli | Tua Rita | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | Medium | High | Low | Medium | High | Low | Medium | High | Low | Medium | High | |
| F3 | 32.15 | 42.41 | 55.0 | 56.1 | 86.1 | 115.61 | 23.56 | 44.47 | 57.62 | 36.9 | 48.96 | 56.05 |
| F4 | 47.13 | 63.2 | 106.2 | 83.22 | 101.71 | 117.25 | 54.85 | 84.08 | 96.96 | 33.81 | 62.6 | 70.61 |
| F5 | 54.78 | 75.25 | 84.36 | 90.8 | 100.49 | 107.85 | 48.03 | 84.04 | 92.27 | 49.92 | 58.01 | 68.31 |
| F6 | 56.77 | 76.06 | 79.2 | 88.64 | 97.14 | 101.62 | 88.46 | 85.31 | 108.53 | 43.25 | 58.58 | 69.22 |
| Casadei | Petra | Rigoli | Tua Rita | |||||
|---|---|---|---|---|---|---|---|---|
| F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | |
| F3 | 2.928 | 0.084 | 0.472 | 0.629 | 15.203 | <0.001 | 0.568 | 0.575 |
| F4 | 2.919 | 0.09 | 0.897 | 0.421 | 2.715 | 0.103 | 1.808 | 0.196 |
| F5 | 0.015 | 0.985 | 0.146 | 0.865 | 5.059 | 0.024 | 3.045 | 0.069 |
| F6 | 0.542 | 0.594 | <0.001 | 1.0 | 0.726 | 0.513 | 2.266 | 0.129 |
| Casadei | Petra | Rigoli | Tua Rita | |||||
|---|---|---|---|---|---|---|---|---|
| F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | |
| F3 | 2.891 | 0.087 | 0.721 | 0.497 | 11.755 | 0.001 | 1.323 | 0.288 |
| F4 | 5.874 | 0.015 | 1.36 | 0.276 | 5.117 | 0.023 | 0.926 | 0.417 |
| F5 | 0.009 | 0.991 | 0.125 | 0.883 | 3.124 | 0.078 | 4.946 | 0.017 |
| F6 | 0.63 | 0.548 | 0.206 | 0.815 | 3.045 | 0.104 | 8.997 | 0.002 |
| Casadei | Petra | Rigoli | Tua Rita | |||||
|---|---|---|---|---|---|---|---|---|
| F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | F Statistic | p Value | |
| F3 | 4.921 | 0.023 | 5.457 | 0.011 | 11.543 | 0.001 | 4.235 | 0.029 |
| F4 | 19.247 | <0.001 | 4.966 | 0.016 | 5.212 | 0.022 | 12.036 | 0.001 |
| F5 | 2.009 | 0.174 | 1.368 | 0.274 | 9.516 | 0.003 | 3.797 | 0.039 |
| F6 | 1.43 | 0.275 | 0.608 | 0.553 | 3.007 | 0.106 | 9.583 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Govi, D.; Pappalardo, S.E.; De Marchi, M.; Meggio, F. From Space to Field: Combining Satellite, UAV and Agronomic Data in an Open-Source Methodology for the Validation of NDVI Maps in Precision Viticulture. Remote Sens. 2024, 16, 735. https://doi.org/10.3390/rs16050735
Govi D, Pappalardo SE, De Marchi M, Meggio F. From Space to Field: Combining Satellite, UAV and Agronomic Data in an Open-Source Methodology for the Validation of NDVI Maps in Precision Viticulture. Remote Sensing. 2024; 16(5):735. https://doi.org/10.3390/rs16050735
Chicago/Turabian StyleGovi, David, Salvatore Eugenio Pappalardo, Massimo De Marchi, and Franco Meggio. 2024. "From Space to Field: Combining Satellite, UAV and Agronomic Data in an Open-Source Methodology for the Validation of NDVI Maps in Precision Viticulture" Remote Sensing 16, no. 5: 735. https://doi.org/10.3390/rs16050735