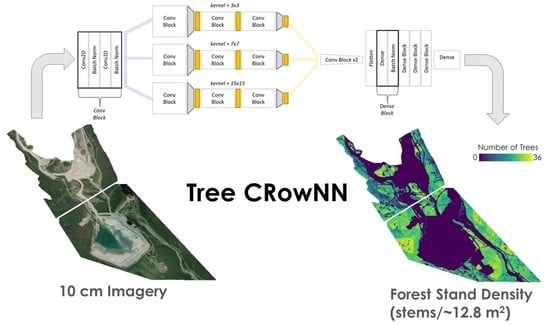
Tree-CRowNN: A Network for Estimating Forest Stand Density from VHR Aerial Imagery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites
- 1.
- Recently harvested with disbursed saplings.
- 2.
- Immature, dense regrowth of trees.
- 3a.
- Mature regrowth of trees with homogenous growth patterns.
- 3b.
- Mature regrowth of trees with heterogeneous growth patterns.
- 4.
- Mature, undisturbed forest.
2.2. Data Preparation
2.3. Model Development
2.4. Model Training
3. Results
3.1. Accuracy Assessment and Performance across Forest Conditions
3.2. Comparison with Sentinel-2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, C.; Zhou, J.; Wang, H.; Tan, T.; Cui, M.; Huang, Z.; Wang, P.; Zhang, L. Multi-Species Individual Tree Segmentation and Identification Based on Improved Mask R-CNN and UAV Imagery in Mixed Forests. Remote Sens. 2022, 14, 874. [Google Scholar] [CrossRef]
- Guang, C.; Shang, Y. Transformer for Tree Counting in Aerial Images. Remote Sens. 2022, 14, 476. [Google Scholar] [CrossRef]
- Marchi, M. Nonlinear versus linearised model on stand density model fitting and stand density index calculation: Analysis of coefficients estimation via simulation. J. For. Res. 2019, 30, 1595–1602. [Google Scholar] [CrossRef]
- Wylie, R.; Woods, M.; Dech, J.P. Estimating Stand Age from Airborne Laser Scanning Data to Improve Models of Black Spruce Wood Density in the Boreal Forest of Ontario. Remote Sens. 2019, 11, 2022. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, D.; Zhou, R.; Fang, L.; Zheng, X.; Lou, X. Estimation of DBH at Forest Stand Level Based on Multi-Parameters and Generalized Regression Neural Network. Forests 2019, 10, 778. [Google Scholar] [CrossRef]
- Fehérváry, I.; Kiss, T. Riparian vegetation density mapping of an extremely densely vegetated confined floodplain. Hydrology 2021, 8, 176. [Google Scholar] [CrossRef]
- Stephens, S.L.; Bernal, A.A.; Collins, B.M.; Finney, M.A.; Lautenberger, C.; Saah, D. Mass fire behavior created by extensive tree mortality and high tree density not predicted by operational fire behavior models in the southern Sierra Nevada. For. Ecol. Manag. 2022, 518, 120258. [Google Scholar] [CrossRef]
- Environment and Climate Change Canada. Government of Canada Adaptation Action Plan (No. 978-0-660-46354–4) 2022, Government of Canada. Available online: https://www.canada.ca/content/dam/eccc/documents/pdf/climate-change/climate-plan/national-adaptation-strategy/GCAAP-Report-EN.pdf (accessed on 27 September 2023).
- Canadian Forestry Service (CFS). Canada’s National Forest Inventory Ground Sampling Guidelines: Specifications for Ongoing Measurement; Version 5; Canadian Forestry Service (CFS): Edmonton, AB, Canada, 2008; ISBN 978-1-100-11329-6. [Google Scholar]
- Bolat, F.; Bulut, S.; Günlü, A.; Ercanli, İ.; Şenyurt, M. Regression kriging to improve basal area and growing stock volume estimation based on remotely sensed data, terrain indices and forest inventory of black pine forests. N. Z. J. For. Sci. 2020, 50, 1–11. [Google Scholar] [CrossRef]
- Abdollahnejad, A.; Panagiotidis, D.; Surový, P. Forest canopy density assessment using different approaches—Review. J. For. Sci. 2017, 63, 107–116. [Google Scholar] [CrossRef]
- Richardson, G.; Leblanc, S.G.; Lovitt, J.; Rajaratnam, K.; Chen, W. Leveraging AI to Estimate Caribou Lichen in UAV Orthomosaics from Ground Photo Datasets. Drones 2021, 5, 99. [Google Scholar] [CrossRef]
- Dersch, S.; Schöttl, A.; Krzystek, P.; Heurich, M. Towards complete tree crown delineation by instance segmentation with mask R–CNN and DETR using UAV-based multispectral imagery and lidar data. ISPRS Open J. Photogramm. Remote Sens. 2023, 8, 100037. [Google Scholar] [CrossRef]
- Tao, S.; Xiuming, S.; Jing, C. Three-Dimensional Imaging of Spinning Space Debris Based on the Narrow-Band Radar. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1041–1045. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhao, P.; Li, Y.; Wang, H.; Yang, Y. Spectrum Interference-Based Two-Level Data Augmentation Method in Deep Learning for Automatic Modulation Classification. Neural Comput. Appl. 2021, 33, 7723–7745. [Google Scholar] [CrossRef]
- Babu Sam, D.; Peri, S.V.; Narayanan Sundararaman, M.; Kamath, A.; Radhakrishnan, V.B. Locate, Size and Count: Accurately Resolving People in Dense Crowds via Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 2739–2751. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wang, L.; Li, T.; Lu, S.; Yin, Z.; Liu, X.; Li, X.; Zheng, W. U-Net-STN: A Novel End-to-End Lake Boundary Prediction Model. Land 2023, 12, 1602. [Google Scholar] [CrossRef]
- Yao, Y.; Jiang, Z.; Zhang, H.; Cai, B.; Meng, G.; Zuo, D. Chimney and Condensing Tower Detection Based on Faster R-CNN in High Resolution Remote Sensing Images. In Proceedings of the 2017 IEEE International Geoscience and Remote Sensing Symposium (IGARSS) 2017, Fort Worth, TX, USA, 23–28 July 2017; pp. 3329–3332. [Google Scholar]
- Han, P.; Ma, C.; Chen, J.; Chen, L.; Bu, S.; Xu, S.; Zhao, Y.; Zhang, C.; Hagino, T. Fast Tree Detection and Counting on UAVs for Sequential Aerial Images with Generating Orthophoto Mosaicing. Remote Sens. 2022, 14, 4113. [Google Scholar] [CrossRef]
- Picos, J.; Bastos, G.; Miguez, D.P.; Alonso, L.C.; Armesto, J. Individual Tree Detection in a Eucalyptus Plantation Using Unmanned Aerial Vehicle (UAV)-LiDAR. Remote Sens. 2020, 12, 885. [Google Scholar] [CrossRef]
- Pleșoianu, A.; Stupariu, M.; Sandric, I.; Pătru-Stupariu, I.; Drăguț, L. Individual Tree-Crown Detection and Species Classification in Very High-Resolution Remote Sensing Imagery Using a Deep Learning Ensemble Model. Remote Sens. 2020, 12, 2426. [Google Scholar] [CrossRef]
- Osco, L.P.; De Arruda, M.D.S.; Junior, J.M.; Da Silva, N.L.; Ramos, A.M.; Moryia, É.A.S.; Imai, N.N.; Pereira, D.R.; Creste, J.E.; Matsubara, E.T.; et al. A convolutional neural network approach for counting and geolocating citrus-trees in UAV multispectral imagery. ISPRS J. Photogramm. Remote Sens. 2020, 160, 97–106. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.A.; Zare, A.; White, E.P. Geographic Generalization in Airborne RGB Deep Learning Tree Detection. bioRxiv 2019. [Google Scholar] [CrossRef]
- Santoso, H.B.; Tani, H.; Wang, X. A simple method for detection and counting of oil palm trees using high-resolution multispectral satellite imagery. Int. J. Remote Sens. 2016, 37, 5122–5134. [Google Scholar] [CrossRef]
- Liu, T.; Yao, L.; Qin, J.; Lu, J.; Liu, N.; Zhou, C. A deep neural network for the estimation of tree density based on High-Spatial Resolution image. IEEE Trans. Geosci. Remote Sens. 2022, 60, 4403811. [Google Scholar] [CrossRef]
- Ocer, N.E.; Kaplan, G.; Erdem, F.; Matci, D.K.; Avdan, U. Tree extraction from multi-scale UAV images using Mask R-CNN with FPN. Remote Sens. Lett. 2020, 11, 847–856. [Google Scholar] [CrossRef]
- Gouiaa, R.; Akhloufi, M.A.; Shahbazi, M. Advances in Convolution Neural Networks Based Crowd Counting and Density Estimation. Big Data Cogn. Comput. 2021, 5, 50. [Google Scholar] [CrossRef]
- Hong, S.; Nam, I.H.; Kim, S.; Kim, E.; Lee, C.; Ahn, S.; Park, I.; Kim, G. Automatic Pest Counting from Pheromone Trap Images Using Deep Learning Object Detectors for Matsucoccus thunbergianae Monitoring. Insects 2021, 12, 342. [Google Scholar] [CrossRef] [PubMed]
- Lempitsky, V.; Zisserman, A. Learning to count objects in images. In Proceedings of the Advances in Neural Information Processing Systems 23 (NIPS 2010), Vancouver, BC, Canada, 6–9 December 2010; p. 23. [Google Scholar]
- Graczyk, K.M.; Pawłowski, J.; Majchrowska, S.; Golan, T. Self-normalized density map (SNDM) for counting microbiological objects. Sci. Rep. 2022, 12, 10583. [Google Scholar] [CrossRef] [PubMed]
- Gao, G. CNN-based Density Estimation and Crowd Counting: A survey. arXiv 2020, arXiv:2003.12783. [Google Scholar] [CrossRef]
- Yao, L.; Liu, T.; Qin, J.; Lu, N.; Zhou, C. Tree counting with high spatial-resolution satellite imagery based on deep neural networks. Ecol. Indic. 2021, 125, 107591. [Google Scholar] [CrossRef]
- Mills, K.; Tamblyn, I. Weakly-supervised multi-class object localization using only object counts as labels. arXiv 2021, arXiv:2102.11743. [Google Scholar]
- Mount Polley Mining Corporation Environmental Department [MPMC]. 2021 Annual Environmental Report for the Mount Polley Mine 2022. Available online: https://www2.gov.bc.ca/gov/content/environment/air-land-water/spills-environmental-emergencies/spill-incidents/past-spill-incidents/mt-polley/mount-polley-key-information (accessed on 31 March 2022).
- Meidinger, D.; Pojar, J. Ecosystems of British Columbia; Ministry of Forests: Vancouver, BC, Canada, 1991; 330p. Available online: https://www.env.gov.bc.ca/thompson/esd/hab/interior_cedar_hemlock.html (accessed on 26 September 2023).
- Huynh, H.X.; Truong, B.C.Q.; Thanh, K.T.N.; Truong, D.Q. Plant Identification Using New Architecture Convolutional Neural Networks Combine with Replacing the Red of Color Channel Image by Vein Morphology Leaf. Vietnam J. Comput. Sci. 2020, 7, 197–208. [Google Scholar] [CrossRef]
- Kattenborn, T.; Eichel, J.; Fassnacht, F.E. Convolutional Neural Networks enable efficient, accurate and fine-grained segmentation of plant species and communities from high-resolution UAV imagery. Sci. Rep. 2019, 9, 17656. [Google Scholar] [CrossRef]
- Rahnemoonfar, M.; Sheppard, C. Deep Count: Fruit Counting Based on Deep Simulated Learning. Sensors 2017, 17, 905. [Google Scholar] [CrossRef] [PubMed]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Zheng, X.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. Available online: https://research.google/pubs/pub45166/ (accessed on 18 February 2020).
- Azizi, Z. Forest canopy density estimating using satellite images. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2008, 8, 1127–1130. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Duchesnay, E. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Macintyre, P.; Van Niekerk, A.; Mucina, L. Efficacy of Multi-Season Sentinel-2 Imagery for Compositional Vegetation Classification. Int. J. Appl. Earth Obs. Geoinf. 2020, 85, 101980. [Google Scholar] [CrossRef]
- Richardson, G.; Knudby, A.; Chen, W. Utilizing Transfer Learning with Artificial Intelligence for Scaling-Up Lichen Coverage Maps. In Proceedings of the IGARSS 2023—2023 IEEE International Geoscience and Remote Sensing Symposium, Pasadena, CA, USA, 16 July 2023; pp. 3050–3053. [Google Scholar]
- Watkins, B.; Van Niekerk, A. A Comparison of Object-Based Image Analysis Approaches for Field Boundary Delineation Using Multi-Temporal Sentinel-2 Imagery. Comput. Electron. Agric. 2019, 158, 294–302. [Google Scholar] [CrossRef]
- Pearse, G.D.; Dash, J.P.; Persson, H.J.; Watt, M.S. Comparison of high-density LiDAR and satellite photogrammetry for forest inventory. ISPRS J. Photogramm. Remote Sens. 2018, 142, 257–267. [Google Scholar] [CrossRef]
- Hashemi, M. Enlarging smaller images before inputting into convolutional neural network: Zero-padding vs. interpolation. J. Big Data 2019, 6, 98. [Google Scholar] [CrossRef]








| Forest Condition | n | MAE | MSE | RMSE |
|---|---|---|---|---|
| 1 | 20 | 2.7 | 12.0 | 3.5 |
| 2 | 20 | 1.8 | 6.0 | 2.4 |
| 3a | 20 | 1.1 | 3.3 | 1.8 |
| 3b | 20 | 2.4 | 14.7 | 3.8 |
| 4 | 20 | 2.5 | 9.8 | 3.1 |
| ALL | 100 | 2.1 | 9.1 | 3.0 |
| Forest Condition | Min | Max | Mean | Median | |
|---|---|---|---|---|---|
| True Counts | 1 | 0 | 30 | 9.2 | 8 |
| 2 | 8 | 26 | 18.7 | 19 | |
| 3a | 0 | 26 | 16.8 | 17 | |
| 3b | 5 | 33 | 16.9 | 17 | |
| 4 | 0 | 18 | 7.5 | 6 | |
| Tree-CRowNN Counts | 1 | 0 | 26 | 7.3 | 3.5 |
| 2 | 5 | 26 | 19.1 | 20 | |
| 3a | 0 | 27 | 17.2 | 18.5 | |
| 3b | 2 | 36 | 16.6 | 18 | |
| 4 | 0 | 19 | 9.8 | 8 |
| Model | Predicted Range | Vs. Tree-CRowNN | ||
|---|---|---|---|---|
| Min | Max | R2 | RMSE | |
| Tree-CRowNN | 1 | 29 | - | - |
| Random Forest | 3 | 24 | 0.433 | 3.9 |
| Linear MSAVI | 5 | 20 | 0.224 | 4.6 |
| Linear NDVI | 5 | 20 | 0.221 | 4.6 |
| Linear NDRE | 6 | 19 | 0.221 | 4.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lovitt, J.; Richardson, G.; Zhang, Y.; Richardson, E. Tree-CRowNN: A Network for Estimating Forest Stand Density from VHR Aerial Imagery. Remote Sens. 2023, 15, 5307. https://doi.org/10.3390/rs15225307
Lovitt J, Richardson G, Zhang Y, Richardson E. Tree-CRowNN: A Network for Estimating Forest Stand Density from VHR Aerial Imagery. Remote Sensing. 2023; 15(22):5307. https://doi.org/10.3390/rs15225307
Chicago/Turabian StyleLovitt, Julie, Galen Richardson, Ying Zhang, and Elisha Richardson. 2023. "Tree-CRowNN: A Network for Estimating Forest Stand Density from VHR Aerial Imagery" Remote Sensing 15, no. 22: 5307. https://doi.org/10.3390/rs15225307