Machine Learning Techniques to Predict Soybean Plant Density Using UAV and Satellite-Based Remote Sensing
Abstract
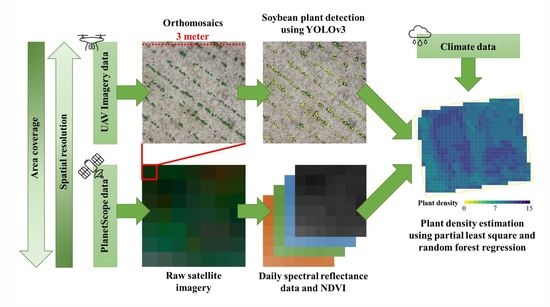
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Imagery and Climate Data Collection
2.2.1. Acquisition of UAV Imagery
2.2.2. PlanetScope Imagery Data
2.2.3. Climate Data
2.3. Actual Plant Density Measurement from UAV Imagery Using YOLOv3
2.4. Daily Spectral Reflectance Gap-Filling of PlanetScope
2.5. Selecting Variables
2.6. PLS and RF Regression
3. Results
3.1. UAV Imagery and YOLOv3 Model Accuracy
3.2. Accuracy of PLS and RF Regression Model
3.3. Spatial Prediction
4. Discussion
4.1. Quality of Measured Actual Plant Density Using YOLOv3 Model
4.2. Factors Affecting the Accuracy of the Regression Models
4.3. Benefit of Variable Importance of Random Forest
4.4. Plant Density Distribution Map and Future Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ball, R.A.; Purcell, L.C.; Vories, E.D. Optimizing Soybean Plant Population for a Short-Season Production System in the Southern USA. Crop Sci. 2000, 40, 757–764. [Google Scholar] [CrossRef]
- Gan, Y.; Stulen, I.; van Keulen, H.; Kuiper, P.J. Physiological response of soybean genotypes to plant density. Field Crops Res. 2002, 74, 231–241. [Google Scholar] [CrossRef]
- De Bruin, J.L.; Pedersen, P. New and Old Soybean Cultivar Responses to Plant Density and Intercepted Light. Crop Sci. 2009, 49, 2225–2232. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Constantin, J.; Schoving, C.; Maury, P.; Debaeke, P.; Aubertot, J.-N.; Dürr, C. Analysis of soybean germination, emergence, and prediction of a possible northward establishment of the crop under climate change. Eur. J. Agron. 2020, 113, 125972. [Google Scholar] [CrossRef]
- Takeda, H.; Sasaki, R. Effects of Ground Water Level Control on the Establishment, Growth and Yield of Soybeans Seeded during and after the Rainy Season. Jpn. J. Crop Sci. 2013, 82, 233–241, (In Japanese with English Summary). [Google Scholar] [CrossRef] [Green Version]
- Carciochi, W.D.; Schwalbert, R.; Andrade, F.H.; Corassa, G.M.; Carter, P.; Gaspar, A.P.; Schmidt, J.; Ciampitti, I.A. Soybean Seed Yield Response to Plant Density by Yield Environment in North America. Agron. J. 2019, 111, 1923–1932. [Google Scholar] [CrossRef] [Green Version]
- Rigsby, B.; Board, J.E. Identification of soybean cultivars that yield well at low plant populations. Crop Sci. 2003, 43, 234–239. [Google Scholar] [CrossRef]
- Egli, D.B. Plant Density and Soybean Yield. Crop Sci. 1988, 28, 977–981. [Google Scholar] [CrossRef]
- Corassa, G.M.; Amado, T.J.C.; Strieder, M.L.; Schwalbert, R.; Pires, J.L.F.; Carter, P.R.; Ciampitti, I.A. Optimum soybean seeding rates by yield environment in southern Brazil. Agron. J. 2018, 110, 2430–2438. [Google Scholar] [CrossRef]
- Gaspar, A.P.; Mourtzinis, S.; Kyle, D.; Galdi, E.; Lindsey, L.E.; Hamman, W.P.; Matcham, E.G.; Kandel, H.J.; Schmitz, P.; Stanley, J.D.; et al. Defining optimal soybean seeding rates and associated risk across North America. Agron. J. 2020, 112, 2103–2114. [Google Scholar] [CrossRef] [Green Version]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Hunt, M.L.; Blackburn, G.A.; Carrasco, L.; Redhead, J.W.; Rowland, C.S. High resolution wheat yield mapping using Sentinel-2. Remote Sens. Environ. 2019, 233, 111410. [Google Scholar] [CrossRef]
- Matese, A.; Toscano, P.; Di Gennaro, S.F.; Genesio, L.; Vaccari, F.P.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B. Intercomparison of UAV, aircraft and satellite remote sensing platforms for precision viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef] [Green Version]
- Dalponte, M.; Frizzera, L.; Gianelle, D. Individual tree crown delineation and tree species classification with hyperspectral and LiDAR data. PeerJ 2019, 2019. [Google Scholar] [CrossRef]
- Wang, J.; Yang, D.; Detto, M.; Nelson, B.W.; Chen, M.; Guan, K.; Wu, S.; Yan, Z.; Wu, J. Multi-scale integration of satellite remote sensing improves characterization of dry-season green-up in an Amazon tropical evergreen forest. Remote Sens. Environ. 2020, 246, 111865. [Google Scholar] [CrossRef]
- Wu, S.; Wang, J.; Yan, Z.; Song, G.; Chen, Y.; Ma, Q.; Deng, M.; Wu, Y.; Zhao, Y.; Guo, Z.; et al. Monitoring tree-crown scale autumn leaf phenology in a temperate forest with an integration of PlanetScope and drone remote sensing observations. ISPRS J. Photogramm. Remote Sens. 2021, 171, 36–48. [Google Scholar] [CrossRef]
- Oh, S.; Chang, A.; Ashapure, A.; Jung, J.; Dube, N.; Maeda, M.; Gonzalez, D.; Landivar, J. Plant counting of cotton from UAS imagery using deep learning-based object detection framework. Remote Sens. 2020, 12, 2981. [Google Scholar] [CrossRef]
- Emilien, A.-V.; Thomas, C.; Thomas, H. UAV & satellite synergies for optical remote sensing applications: A literature review. Sci. Remote Sens. 2021, 3, 100019. [Google Scholar] [CrossRef]
- Blázquez-Casado, Á.; Calama, R.; Valbuena, M.; Vergarechea, M.; Rodríguez, F. Combining low-density LiDAR and satellite images to discriminate species in mixed Mediterranean forest. Ann. For. Sci. 2019, 76. [Google Scholar] [CrossRef]
- Saeys, W.; Lenaerts, B.; Craessaerts, G.; De Baerdemaeker, J. Estimation of the crop density of small grains using LiDAR sensors. Biosyst. Eng. 2009, 102, 22–30. [Google Scholar] [CrossRef]
- Floreano, D.; Wood, R.J. Science, technology and the future of small autonomous drones. Nature 2015, 521, 460–466. [Google Scholar] [CrossRef] [Green Version]
- Ranđelović, P.; Đorđević, V.; Milić, S.; Balešević-Tubić, S.; Petrović, K.; Miladinović, J.; Đukić, V. Prediction of Soybean Plant Density Using a Machine Learning Model and Vegetation Indices Extracted from RGB Images Taken with a UAV. Agronomy 2020, 10, 1108. [Google Scholar] [CrossRef]
- Wu, J.; Yang, G.; Yang, X.; Xu, B.; Han, L.; Zhu, Y. Automatic Counting of in situ Rice Seedlings from UAV Images Based on a Deep Fully Convolutional Neural Network. Remote Sens. 2019, 11, 691. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Liu, S.; Baret, F.; Hemerlé, M.; Comar, A. Estimates of plant density of wheat crops at emergence from very low altitude UAV imagery. Remote Sens. Environ. 2017, 198, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Zan, X.; Zhang, X.; Xing, Z.; Liu, W.; Zhang, X.; Su, W.; Liu, Z.; Zhao, Y.; Li, S. Automatic Detection of Maize Tassels from UAV Images by Combining Random Forest Classifier and VGG16. Remote Sens. 2020, 12, 3049. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, B.; Yang, C.; Shi, Y.; Liao, Q.; Zhou, G.; Wang, C.; Xie, T.; Jiang, Z.; Zhang, D.; et al. Rapeseed Stand Count Estimation at Leaf Development Stages with UAV Imagery and Convolutional Neural Networks. Front. Plant Sci. 2020, 11, 617. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, C.; Paterson, A.H.; Robertson, J.S. DeepSeedling: Deep convolutional network and Kalman filter for plant seedling detection and counting in the field. Plant Methods 2019, 15, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peña, J.M.; Torres-Sánchez, J.; Serrano-Pérez, A.; de Castro, A.I.; López-Granados, F. Quantifying efficacy and limits of unmanned aerial vehicle (UAV) technology for weed seedling detection as affected by sensor resolution. Sensors 2015, 15, 5609–5626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Vrieling, A.; Fava, F.; Meroni, M.; Marshall, M.; Gachoki, S. Phenology of short vegetation cycles in a Kenyan rangeland from PlanetScope and Sentinel-2. Remote Sens. Environ. 2020, 248, 112004. [Google Scholar] [CrossRef]
- Khaliliaqdam, N.; Soltani, A.; Latifi, N.; Ghaderi Far, F. Soybean Seed Aging and Environmental Factors on Seedling Growth. Commun. Soil Sci. Plant Anal. 2013, 44, 1786–1799. [Google Scholar] [CrossRef]
- Bajgain, R.; Kawasaki, Y.; Akamatsu, Y.; Tanaka, Y.; Kawamura, H.; Katsura, K.; Shiraiwa, T. Biomass production and yield of soybean grown under converted paddy fields with excess water during the early growth stage. Field Crops Res. 2015, 180, 221–227. [Google Scholar] [CrossRef]
- Liu, J.; Sun, S.; Tan, Z.; Liu, Y. Nondestructive detection of sunset yellow in cream based on near-infrared spectroscopy and interval random forest. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 242, 118718. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Sakaiya, E.; Zhu, Y.; Takahashi, W. Diagnostic mapping of canopy nitrogen content in rice based on hyperspectral measurements. Remote Sens. Environ. 2012, 126, 210–221. [Google Scholar] [CrossRef]
- Vaglio Laurin, G.; Chen, Q.; Lindsell, J.A.; Coomes, D.A.; Del Frate, F.; Guerriero, L.; Pirotti, F.; Valentini, R. Above ground biomass estimation in an African tropical forest with lidar and hyperspectral data. ISPRS J. Photogramm. Remote Sens. 2014, 89, 49–58. [Google Scholar] [CrossRef]
- Otgonbayar, M.; Atzberger, C.; Chambers, J.; Damdinsuren, A. Mapping pasture biomass in Mongolia using Partial Least Squares, Random Forest regression and Landsat 8 imagery. Int. J. Remote Sens. 2019, 40, 3204–3226. [Google Scholar] [CrossRef]
- Reddy, N.; Gebreslasie, M.; Ismail, R. A hybrid partial least squares and random forest approach to modelling forest structural attributes using multispectral remote sensing data. S. Afr. J. Geomat. 2017, 6, 377. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Gu, S.; Shen, M.; Tang, Y.; Matsushita, B. Estimating aboveground biomass of grassland having a high canopy cover: An exploratory analysis of in situ hyperspectral data. Int. J. Remote Sens. 2009, 30, 6497–6517. [Google Scholar] [CrossRef]
- Matsuo, N.; Yamada, T.; Takada, Y.; Fukami, K.; Hajika, M. Effect of plant density on growth and yield of new soybean genotypes grown under early planting condition in southwestern Japan. Plant Prod. Sci. 2018, 21, 16–25. [Google Scholar] [CrossRef] [Green Version]
- Planet Labs Inc. Planet Imagery Product Specifications. 2021. Available online: https://assets.planet.com/docs/Planet_Combined_Imagery_Product_Specs_letter_screen.pdf (accessed on 19 March 2021).
- Ohno, H.; Sasaki, K.; Ohara, G.; Nakazono, K. Development of grid square air temperature and precipitation data compiled from observed, forecasted, and climatic normal data. Clim. Biosph. 2016, 16, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1312.6229. [Google Scholar]
- Huang, R.; Gu, J.; Sun, X.; Hou, Y.; Uddin, S. A rapid recognition method for electronic components based on the improved YOLO-V3 network. Electronics 2019, 8, 825. [Google Scholar] [CrossRef] [Green Version]
- Tzutalin. LabelImg. Git Code. 2015. Available online: https://github.com/tzutalin/labelImg (accessed on 22 January 2020).
- Rowlands, C.J.; Elliott, S.R. Denoising of spectra with no user input: A spline-smoothing algorithm. J. Raman Spectrosc. 2011, 42, 370–376. [Google Scholar] [CrossRef]
- Rouse, J.W.; Hass, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the great plains with ERTS. In Proceedings of the Third Earth Resources Technology Satellite (ERTS) Symposium, Washington, DC, USA, 10–14 December 1973; Volume 1, pp. 309–317. [Google Scholar]
- Summers, D.; Lewis, M.; Ostendorf, B.; Chittleborough, D. Visible near-infrared reflectance spectroscopy as a predictive indicator of soil properties. Ecol. Indic. 2011, 11, 123–131. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Castaldi, F.; Casa, R.; Castrignanò, A.; Pascucci, S.; Palombo, A.; Pignatti, S. Estimation of soil properties at the field scale from satellite data: A comparison between spatial and non-spatial techniques. Eur. J. Soil Sci. 2014, 65, 842–851. [Google Scholar] [CrossRef]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Mevik, B.-H.; Wehrens, R. Introduction to the pls Package. 2015. Available online: https://cran.r-project.org/web/packages/pls/vignettes/pls-manual.pdf (accessed on 28 December 2020).
- Breiman, L. Random forests. Mach. Learn. 2001. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M.; Breimann, L.; Cutler, A. Randomforest: Breiman and Cutler’s Random Forests for Classification and Regression. 2018. Available online: https://cran.r-project.org/web/packages/randomForest/randomForest.pdf (accessed on 15 January 2021).
- Strobl, C.; Boulesteix, A.L.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional variable importance for random forests. BMC Bioinform. 2008, 9, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, C.; Mira-McWilliams, J.; Juárez, I. Important variable assessment and electricity price forecasting based on regression tree models: Classification and regression trees, Bagging and Random Forests. IET Gener. Transm. Distrib. 2015, 9, 1120–1128. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, G.; Wang, J.; Song, X.; Feng, H. Winter wheat biomass estimation based on spectral indices, band depth analysis and partial least squares regression using hyperspectral measurements. Comput. Electron. Agric. 2014, 100, 51–59. [Google Scholar] [CrossRef]
- Cho, M.A.; Skidmore, A.; Corsi, F.; van Wieren, S.E.; Sobhan, I. Estimation of green grass/herb biomass from airborne hyperspectral imagery using spectral indices and partial least squares regression. Int. J. Appl. Earth Obs. Geoinf. 2007, 9, 414–424. [Google Scholar] [CrossRef]
- Testa, S.; Soudani, K.; Boschetti, L.; Borgogno Mondino, E. MODIS-derived EVI, NDVI and WDRVI time series to estimate phenological metrics in French deciduous forests. Int. J. Appl. Earth Obs. Geoinf. 2018, 64, 132–144. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Hyperspectral band depth analysis for a better estimation of grass biomass (Cenchrus ciliaris) measured under controlled laboratory conditions. Int. J. Appl. Earth Obs. Geoinf. 2004, 5, 87–96. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, M.A. High density biomass estimation for wetland vegetation using worldview-2 imagery and random forest regression algorithm. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Zhou, X.; Kono, Y.; Win, A.; Matsui, T.; Tanaka, T.S.T. Predicting within-field variability in grain yield and protein content of winter wheat using UAV-based multispectral imagery and machine learning approaches. Plant Prod. Sci. 2020, 24, 1–15. [Google Scholar] [CrossRef]
- Strobl, C.; Boulesteix, A.L.; Zeileis, A.; Hothorn, T. Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinform. 2007, 8, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawasaki, Y.; Yamazaki, R.; Katayama, K. Effects of late sowing on soybean yields and yield components in southwestern Japan. Plant Prod. Sci. 2018, 21, 339–348. [Google Scholar] [CrossRef] [Green Version]
- Hodges, T.; French, V. Soyphen: Soybean Growth Stages Modeled from Temperature, Daylength, and Water Availability. Agron. J. 1985, 77, 500–505. [Google Scholar] [CrossRef]











| Field Name | Area (ha) | Latitude | Longitude | Sowing Dates | UAV Imagery Acquisition Date | The Number of PlanetScope Imagery a |
|---|---|---|---|---|---|---|
| Field 1 | 0.9 | 35°11′13.2″ | 136°40′4.8″ | Jul 15, 2018 | 6 August 2018 (22 DAS) | 12 |
| Field 2 | 1.16 | 35°11′31.2″ | 136°40′1.2″ | Jul 15, 2018 | 6 August 2018 (22 DAS) | 12 |
| Field 3 | 1.1 | 35°11′20.4″ | 136°39′54.0″ | Jul 17, 2019 | 29 July 2019 (12 DAS) | 13 |
| Field 4 | 1.19 | 35°11′20.4″ | 136°39′54.0″ | Jul 17, 2019 | 29 July 2019 (12 DAS) | 13 |
| Field 5 | 1.11 | 35°11′16.8″ | 136°39′54.0″ | Jul 10, 2019 | 30 July 2019 (20 DAS) | 11 |
| Field 6 | 0.94 | 35°11′16.8″ | 136°39′54.0″ | Jul 10, 2019 | 30 July 2019 (20 DAS) | 11 |
| Field 7 | 0.88 | 35°11′2.4″ | 136°38′16.8″ | Jul 25, 2019 | 5 August 2019 (11 DAS) | 10 |
| Field 8 | 0.95 | 35°11′2.4″ | 136°38′9.6″ | Jul 25, 2019 | 5 August 2020 (11 DAS) | 10 |
| Field 9 | 1.87 | 35°11′16.8″ | 136°39′43.2″ | Aug 2, 2020 | 19 August 2020 (17 DAS) | 8 |
| Field 10 | 0.73 | 35°14′42.0″ | 136°39′50.4″ | Jul 21, 2020 | 4 August 2020 (14 DAS) | 6 |
| Field 11 | 0.61 | 35°14′27.6″ | 136°40′1.2″ | Aug 5, 2020 | 18 August 2020 (13 DAS) | 8 |
| Field 12 | 0.76 | 35°10′4.8″ | 136°39′36.0″ | Aug 4, 2020 | 18 August 2020 (14 DAS) | 8 |
| Variable Type | Dataset |
|---|---|
| Reflectance values | PlanetScope surface reflectance: blue, green, red, NIR at 10, 20, 30, and 40 DAS) |
| Vegetation indices | Normalized difference vegetation index (NDVI) at 10, 20, 30, and 40 DAS |
| Climate data | Precipitation data: seven-days before and after the sowing date |
| Ten-days cumulative temperature at 10, 20, 30, and 40 DAS |
| Model Combination a | PLS Regression | RF Regression | ||||||
|---|---|---|---|---|---|---|---|---|
| Training | Test | Training | Test | |||||
| r2 | RMSE | r2 | RMSE | r2 | RMSE | r2 | RMSE | |
| SR only | 0.71 | 1.81 | 0.50 | 2.17 | 0.84 | 1.37 | 0.63 | 1.82 |
| NDVI only | 0.58 | 2.20 | 0.56 | 2.37 | 0.79 | 1.54 | 0.60 | 1.92 |
| SR + NDVI | 0.74 | 1.74 | 0.56 | 2.08 | 0.84 | 1.35 | 0.67 | 1.81 |
| SR + Climate | 0.76 | 1.65 | 0.19 | 3.67 | 0.84 | 1.36 | 0.63 | 1.83 |
| NDVI + Climate | 0.74 | 1.72 | 0.46 | 2.22 | 0.81 | 1.49 | 0.67 | 1.78 |
| SR + NDVI + Climate | 0.77 | 1.62 | 0.28 | 2.95 | 0.84 | 1.35 | 0.65 | 1.79 |
| Test Field | RMSE (plant m−2) | ||
|---|---|---|---|
| SR + NDVI-PLS | NDVI + Climate-RF | 10VarImp-RF | |
| Field 2 | 1.60 | 1.38 | 1.49 |
| Field 3 | 2.46 | 2.43 | 2.28 |
| Field 8 | 3.06 | 1.90 | 1.82 |
| Field 9 | 1.49 | 1.39 | 1.38 |
| Field 11 | 1.84 | 1.93 | 1.74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habibi, L.N.; Watanabe, T.; Matsui, T.; Tanaka, T.S.T. Machine Learning Techniques to Predict Soybean Plant Density Using UAV and Satellite-Based Remote Sensing. Remote Sens. 2021, 13, 2548. https://doi.org/10.3390/rs13132548
Habibi LN, Watanabe T, Matsui T, Tanaka TST. Machine Learning Techniques to Predict Soybean Plant Density Using UAV and Satellite-Based Remote Sensing. Remote Sensing. 2021; 13(13):2548. https://doi.org/10.3390/rs13132548
Chicago/Turabian StyleHabibi, Luthfan Nur, Tomoya Watanabe, Tsutomu Matsui, and Takashi S. T. Tanaka. 2021. "Machine Learning Techniques to Predict Soybean Plant Density Using UAV and Satellite-Based Remote Sensing" Remote Sensing 13, no. 13: 2548. https://doi.org/10.3390/rs13132548