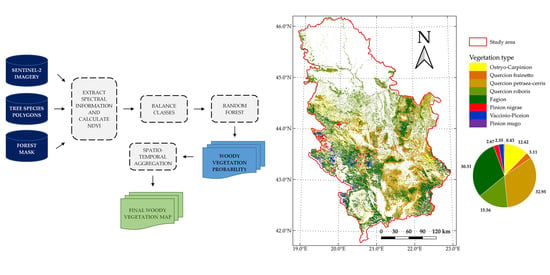
5.1. Final Classification Map of Woody Vegetation
The previously explained LLOCV procedure can give information about the model accuracy, but the accuracy metrics are generally biased for mapping the entire study area. Unfortunately, no comparable external data source is available for the purpose of assessing the accuracy of the final woody vegetation map. In addition, creating a validation set based on probability sampling requires extensive resources and field work, which are beyond the scope of this research. Therefore, validation of the final classification map of woody vegetation has been performed by using visual inspection and comparison with available statistical reports.
By visual inspection and comparison, the final classification map of woody vegetation produced in this paper corresponds to the existing general vegetation maps: Map of the Natural Vegetation of Europe (scale 1:2,500,000) [
29] and the Map of Natural Potential Vegetation of SFR Yugoslavia (scale 1:1,000,000) [
45]. Additionally, the proportional relationships of the analyzed vegetation types for most of the classes fit into the results obtained by the first National Forest Inventory of Serbia in the period 2004–2006, which was done using traditional field survey methods that provide a highly accurate forest inventory [
46,
47]. Significant deviations were obtained only in the proportions of thermophytic broadleaved deciduous forests and mesophytic and hygromesophytic broadleaved forests. The share of Ostryo-Carpinion forests in this research is 12.42%, compared to the share of
Carpinus orientalis and
Ostrya carpinifolia forests in [
47], which is 3.9%. The results of this research show that Quercion roboris forests cover 16% of the area, while the corresponding forest stand category in [
47]
Quercus robur forests has the coverage of 1.4%.
In the first case (Ostryo-Carpinion), the discrepancy can be explained by the fact that the data from the National Forest Inventory of Serbia refer primarily to forests and do not include data on shrubs and lower degradation forms of forest vegetation. Since in the thermophilic forest zone, the degradation forms of forest vegetation are mainly represented by shrub vegetation belonging to the Ostryo-Carpinion type, the increased share of this type of vegetation in our research can be explained by the assumption that different degradation forms of thermophytic broadleaved were recognized in this class. In the second case (Quercion roboris), the deviation is explained by the fact that our class Quercion roboris is much wider than the class
Quercus robur forests presented in the first National Forest Inventory of Serbia (
Appendix A).
Since the spatial distribution of the training polygons cannot be considered ideal, it is possible that some discrepancies in the area assessments are also caused as a result. Therefore, additional woody vegetation polygons in the forest areas sparsely covered by polygons (like southern part of the study area) can significantly benefit in better modeling classes in RF model and more accurate area assessment.
5.3. Spatio-Temporal Framework for Woody Vegetation Classification
The spatio-temporal framework proposed in this research defines the timestamp of the observation explicitly in the model through day and month variables. Such change in the model means that, in order to determine the woody vegetation class, the methodology requires only one cloud-free pixel available for each part of the study area. This change in the model allows timestamps to be freely combined and enables the entire methodology to be easily adjusted to any area of interest, regardless of its size. Additionally, the model is lighter, as every spectral band is represented only once through a single variable, as an alternative to multiple timestamp–spectral band variable combinations. There is also no data-wrangling and only the original observations are used for both training and prediction phases. As the number of cloud-free pixels over the study area increases, there will be multiple predictions for each pixel of the study area. This enables the full potential of spatio-temporal aggregation to be exploited. The spatio-temporal aggregation is not only pixel-based, but is expanded to take into account the neighboring pixel predictions as well. The main idea behind the neighborhood-based approach is to solve situations where surrounding pixels show strong probability of belonging to a certain class, which is different from the class probability of the central pixel. In these cases, there is a strong reason to believe that the central pixel should also belong to the class of surrounding pixels. It is more likely that the prediction based solely on spectral information for the central pixel is wrong, which is a consequence of some noise present in the spectral information for that pixel, than the probability that the central pixel’s class differs from the class of its neighbors.
Applied spatio-temporal aggregation can be affected by temporal and/or spatial homogeneity. For areas with no temporal and spatial heterogeneity over the time period covered with satellite observations and the used neighborhoods, the prediction results should be very reliable. In other cases, the proposed methodology can deal with such heterogeneity up to a certain amount. This is because each of the used S2 observation influences the decision on the final woody vegetation class and any temporal or spatial change can potentially suppress other observations. Temporally, this means that changes that last longer within the used time interval are more likely to be recognized by the model. Similarly, only spatial changes that have larger spatial extents compared to the neighborhood size will be recognized. This means that the minimum mapping unit gets coarser as the size of neighborhood increases. At the same time, the neighborhood approach achieves smoothing effects, where isolated pixels are classified into the class of pixels in the neighborhood. Therefore, it is not advised to extend both the observation period and the neighborhood size more than necessary. A shorter observation period reduces the amount of changes that occur in the first place, and the appropriate neighborhood size limits the amount of smoothing which occurs.
The proposed methodology is applied using resampled 60 m Sentinel-2 bands. This spatial resolution may be too coarse, since the increase in neighborhood size has a limited effect on the performance. It is expected that using the original spatial resolution of 10 m and 20 m for available bands, should reduce the effects of mixed pixels and further increase the classification accuracy. In addition, this should help the previously discussed heterogeneity issues, where smaller changes could be modelled. Stronger neighborhood effects can also be expected to occur for higher spatial resolutions, so the neighborhood size could become a more important parameter in the proposed methodology. However, there are several motivations for choosing a coarser spatial resolution of 60 m for all bands. Firstly, this is done in order to match the chosen classification scheme, designed for country-wide woody vegetation mapping. The used classification scheme is defined according to the “Database on the distribution of potentially endangered species and habitats of Serbia” and the increase in spatial resolution would require the introduction of new woody vegetation classes for which the proper training data are lacking. Secondly and more importantly, the proposed methodology aims to be used on large areas and this way the computation and performance issues are reduced. At a spatial resolution of 60 m, the used dataset comprises more than 160 million S2 observations, which would increase about nine or 36 times in the case of 20 m and 10 m spatial resolutions, respectively. One solution to deal with such an increase in size of data is to select only more informative spectral bands. Since the proposed methodology has advantages and applicability, especially over larger areas, this can become a very important issue.
The results of this research match those obtained by other similar studies, which were based on high- or medium-resolution optical data. Grabska et al. have achieved an OA greater than 90% on the study area of 305 km
2 for nine tree species using 18 cloud-free sentinel-2 images [
6]. Liu et al. identified forest types using Sentinel-1A, Sentinel-2A, Landsat-8 and DEM Data with OA of 82.78%. They extracted the four single-dominant forests and the four mixed forest types over the area of 2261 km
2 [
40]. Lu et al. implemented Spatial–Spectral–Temporal Integrated Fusion to identify six tree species and two mixed forest types with an OA of 83.6% over the area of 1610 km
2 [
49]. In addition to covering a significantly larger study area, this research and the proposed solution also completely relies only on optical satellite sources. Therefore, there is a potential for further improvement by using other data sources such as radar satellite data, LiDAR, multiple optical sources and/or other auxiliary data (topography, etc.).
This proposed spatio-temporal framework relies on Sentinel-2 scene classification maps and a forest mask to properly identify forest areas. This is potentially the weak point of the method, because the output quality is in a way limited by the quality of the used forest mask. It is important to use the most recent available forest mask as possible, especially in the areas of rapid forest change. The forest mask used in this research is from 2015, which creates a 4-year gap with the used Sentinel-2 data. Such a forest mask can be considered outdated since there is an active forest management over the study area, with the forest area being influenced by felling of trees, forest growing and silviculture. Additionally, some forest fires occur annually, usually in the warm summer period. However, these changes are not extensive and do not have large spatial extents. Visual inspection of forest mask showed that the misrepresentations happen only in a few local areas, so the authors believe that these errors do not significantly affect the overall results. The methodology can also be further expanded to address this issue, with the same spatio-temporal framework being used to firstly identify the forest and non-forest pixels. This issue was beyond the scope of this research due to the lack of proper training data for good representation of non-forest areas. This issue needs to be addressed in the future. Another setback of the proposed approach is that each pixel is always assigned to some class. In a situation where all woody vegetation types are well represented by training polygons, this does not pose a problem; however, when the study area is very complex or insufficiently researched in terms of woody vegetation, it might not be possible to represent all the woody vegetation types in the study area.
The study has revealed several problematic cases in which predictions do not achieve a satisfactory level of accuracy and reliability. Some of them are problems with: (1) identification of sufficiently representative learning polygons within the complex and mosaic area, such as alluvial forests in riparian and swamp galleries and woodland in alluvial plains; (2) insufficiently researched natural vegetation types, such as forests of birch (Betula pendula), common aspen (Populus tremula) or rowan (Sorbus aucuparia); (3) the natural absence of clear boundaries between ecologically and physiognomically close vegetation types that build phylogenetically close tree species (e.g., gradual transition between thermophytic (Quercion frainetto), thermo-mesophytic (Quercion petraea-cerris) and mesophytic to hygromesophytic (Quercion roboris) forests in which all three main oak species have such a wide ecological valence that their populations occur within all three types of oak forests); (4) a priori classification of mixed deciduous and coniferous woodlands in which a large number of tree species have different parts in the general coverage; (5) an absence of data on areas with highly artificial tree plantations or naturalized forest of alien species(e.g., black locust forests (Robinia pseudoaccacia), american ash (Fraxinus spp.) and maple (Acer spp.) forests, poplar plantations (Populus spp.), etc.
Given that eight analyzed woody vegetation classes cover a large portion of the study area, it can be expected that the omission of other classes should not significantly affect the spatial coverage of woody vegetation in the study area. Since there are multiple predictions for each pixel of the study area, as well as the probability of each prediction, there is a possibility to use this information to identify the weak pixels, which probably do not belong to any of the classes. This is also something to be considered in future research.