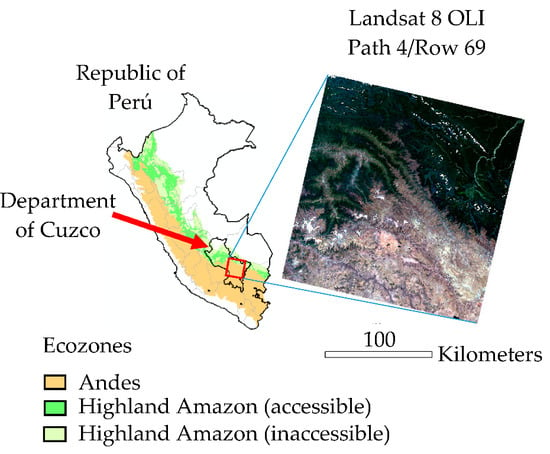
2.3.1. Preprocessing
Figure 2 shows the flow of analysis. One problem with land cover mapping using remote sensing in mountainous terrain is the presence of sunlit and shadowed slopes produced by the steep topography. Topographic correction is necessary to reduce this effect.
After conducting atmospheric and topographic correction for the scenes (P2) and masking of clouds, cloud shadows, and pixels with overcorrected reflectance (P3 and P4), we normalized the satellite data. We conducted this process on pairs of overlapping scenes using the Iteratively Reweighted Multivariate Alteration Detection (IR-MAD) algorithm (P5) [
21,
22] to find pseudo-invariant features that can compute a modified model [
23]. The size of sample areas was 10 km × 10 km, and the sample areas included deep water bodies, concrete, and/or bare soil, aside from forest areas. This method is based on canonical correlation analysis (CCA) and is an unsupervised change detection algorithm that is invariant to linear transformations of the original data. In this method, CCA creates linear combinations of the pixel values for each of the spectral bands of the two scenes. Each pair of linear combinations are called canonical variates (CVs), and the number of CVs is equal to the number of bands. The first pair of CVs has maximal correlation, the second pair of CVs has the second highest correlation and is orthogonal to the first pair of CVs, and so on. Then, the process to determine the difference between the CVs is carried out to produce a sequence of transformed difference images called MAD variates that records the maximum spread (or maximum change) of the pixel values. The sequence generates the same number of MAD variates as the number of bands used. From these different images, we select all pixels with minimum or no change (called Pseudo-Invariant Features, PIFs) that satisfy Formula (1):
where
N is the number of MAD variates,
σ is the variance of the no-change distribution, and
t is a decision threshold value. In the absence of change, the sum of the squares of standardized MAD variates is approximately chi-squared distributed with
N degrees of freedom. The value of
t is defined as:
where
P is the probability of observing a value lower than
t. The process uses
P as weight for the observations, and the whole process is iterated until a stopping criterion is met. In this study, we used a fixed number of iterations equal to 50. With this process, we selected PIFs to carry out linear regressions to produce linear equations for each band to normalize the reflectance values of the scenes. Finally, we created cloud-free normalized mosaics by overlapping the normalized scenes.
In contrast to pixel-based and unsupervised classification techniques, object-based image classification creates land cover maps that are easier to compare with reality. The object-based classification approach implies the creation of “objects” or “groups of pixels.” For this purpose, we carried out a series of a segmentation analyses using eCognition [
24]. This process uses a region-merging algorithm starting with randomly selected pixel seeds that are distributed regularly [
25]. The pixels are then grouped with other pixels based on the homogeneity criteria to form polygons called objects. By using a series of tests with different scale values and evaluating changes in the average size of the created objects, we determined an optimum segmentation scale value to produce objects with an average area equal to the minimum defined forest area (2 ha). Then, we segmented the cloud-free normalized mosaic with the optimum scale value (P6) to produce the objects we used in the classification analysis.
2.3.3. Classification Variables and Variable Selection
The values of the variables used in this study were obtained by an object-based analysis, which means that we used the values of the pixels within each object to calculate the value of the corresponding variable for each object and used the average value in our analyses. The raster datasets used were the cloud-free Landsat 8 OLI mosaic and STRM 30-m DEM data described in
Table 1. Using the longitude and latitude of the land cover data records in the training and verification datasets, we determined the objects corresponding to each land cover point and then extracted the calculated value of the variables from those objects using the statistical software R version 3.4 (
http://www.cran. R-project.org).
The average reflectance values of the visible bands (blue, green, and red), the near infrared (NIR) band, and two shortwave infrared bands were derived from the spectral data. Additionally, we used the three first principal components produced by principal component analysis and tasseled cap transformation, both of which reduce data dimensionality, taking into account the variability of the data as much as possible. We also used the Normalized Difference Vegetation Index (NDVI) and the Modified Soil-adjusted Vegetation Index (MSAVI). MSAVI was calculated as:
where
ρNIR and
ρred are the reflectance values in the NIR and red bands, respectively. This index has been used in vegetation studies in arid and semi-arid regions [
26,
27,
28] because it reduces the soil background effect. We chose to use it among other soil-adjusted vegetation indexes because it can be used without any preliminary knowledge of the vegetation cover rate [
29].
We included elevation and aspect (the downward direction of the slope in degrees) calculated from the DEM data as topography-derived variables in the classification analysis, considering the ecological relationships between vegetation species, altitude, and slope orientation.
One of the characteristics of SVM and RF algorithms is that they do not require feature selection [
30]. However, the performance of classifiers produced by kNN algorithm is affected by the presence of irrelevant or redundant features in the training data [
31,
32]. Since we wanted to determine which among these three machine learning algorithms can produce the most accurate classifier for Andes mountain forest classification, we chose to make such a comparison using the same models produced after feature selection and using the same subsets of data produced with cross validation. Additionally, in order to take into account the full capacity of the SVM and RF algorithms to produce highly accurate classifiers, we introduced and tested classifiers produced with all the available variables.
The variables were subjected to a selection process involving two analyses. In the first analysis, we determined the relative importance of the variables by using the Akaike weight [
33], which is based on the Akaike information criterion (AIC), taking into account that our classification analysis had a binary outcome: Andes mountain forest or shrubland. To do this, we built models using all possible combination of the variables and calculated their corresponding AIC values. Then, we calculated the AIC difference (Δ
i) between each model (
AICi) and the model with minimum AIC (
AICmin):
If
R is the total number of models, then the Akaike weight (
wi) can be calculated for each model as follows:
The relative importance of each variable was estimated by summing the Akaike weights of all the models in which the variable occurs. In the second analysis, we calculated the Pearson’s correlation coefficient between each pair of variables. We chose the variables with the higher values of relative importance and discarded the correlated variables with the lower importance value.
2.3.4. Classification Methods
RF is a powerful MLA that is widely used to classify imagery data for land cover classification using multispectral satellite sensor imagery. The method performs well when the number of predictors is greater than the number of observations and has low sensitivity when the number of irrelevant predictors is large. SVM is another MLA used for classification to determine a hyperplane (or boundary in a high dimensional space) that can divide training data into a predetermined number of categories. This method is used in many remote sensing studies because of its capacity to process small training datasets [
34]. The kNN method is simple to implement and has a low training computational cost. This non-parametric method uses the
k closest training data vectors to make predictions and has been used in forest inventory practices and as a tool for forest classification and mapping [
35,
36,
37].
We carried out a comparative statistical analysis of the three MLAs using non-parametric statistical tests to compare the performances of all the considered models.
1. Random Forest
The RF method [
38] is an ensemble of classification trees in which each tree contributes a unit vote to determine the most frequent class according to the input data (Equation (6)):
where
is the predicted class from the RF classification of data record
x, and
is the predicted class from the classification tree
mi of the data record
x. Each classification tree is constructed using a bootstrap sample of 63.2% of the training data, while the rest of the data is considered out-of-bag (OOB) data. When forming a split point (node) in a tree, the algorithm randomly selects a sub-set of variables and searches among these variables for the best split point to classify the data. The number of variables in each sub-set is commonly denoted as
mtry. The performance of this algorithm depends on the availability of a sufficient number of trees (
ntrees) to be generated to converge on the value of the OOB error and the number of variables randomly sampled as candidate variables in each node of the classification trees (
mtry).
We used 4000 random decision trees. The OOB error is the average of the misclassification(s) rates computed from each sample of the OOB data when classified by all the trees constructed without such samples. We adjusted the value of mtry by carrying out a series of classification tests with different values of this parameter and calculated the corresponding Cohen’s kappa value. Then, we selected the value of mtry that had the maximum Cohen’s kappa value.
2. Support Vector Machine
SVM [
39] is a non-parametric supervised statistical learning classifier that finds a hyperplane for optimal classification by minimizing the upper bound of the classification error. To use this method, we standardized the values of the variables in the training data. The method maximizes the distance from the data points of two classes (in the case of binary classification) to an optimal separation vector of a hyperplane created from the variables [
40]. The hyperplane is the surface used to determine the classification.
Given a training set (
xi,
yi), where
yi is the class label that takes the value of –1 or 1 and
xi is the training vector of the values of the corresponding explanatory variables, a solution to the following optimization problem is needed:
where
φ is a projection function of the training vector given by the kernel model used by the analyst;
w and
b are the adjustable weight and bias parameters, respectively;
C is the penalty parameter of the error term
ξ;
l is the number of samples in the training dataset; and
T denotes the transpose operator.
The parameters
w and
b in Equation (8) define the decision hyper-plane that separates the classes, and the minimization of Equation (7) aims to maximize the separation margin of the data. The projection function is related to a kernel function
K by the following expression:
We used the radial basis function kernel that depends on the value of the parameter
γ in the following expression:
where
xi and
xi’ are two different training vectors of the values of the explanatory variables;
xij and
xi’j are the values of the
jth explanatory variable in the
ith and
i’th training vectors; and
p is the total number of variables. The parameter
γ defines the extent to which the impact of a single training example extends to determine the decision surface. On the other hand,
C trades off misclassification of training data against the number of dimensions that the decision surface should have. These two parameters were adjusted using a grid search analysis; that is, the best decision hyper-plane of the largest Cohen’s kappa value was calculated using different values of
C and
γ in a series of sequential tests.
3.k-Nearest Neighbors
kNN is a well-known nonparametric classification method that assigns a sample vector
x to the class represented by the majority of
k nearest neighbors whose similarity is determined by the distance measure. As with the SVM algorithm, the values of the variables in the training data need to be standardized to use this method. We used the Minkowski metric to define distance:
where
xi is the predictor vector of length
p of observation
i to be classified, and
xj is the
jth nearest neighbor. Euclidean distances can be determined by setting the value of
q = 2. When
kr is the number of nearest neighbors to the observation
xi that belongs to class
r, then:
The algorithm assigns observation xi to the class r for which kr is the largest. We restricted the value of k to odd values to avoid the possibility of a tie between the numbers of neighboring training data samples of two different classes. Because the performance of this method depends on the value of k, we adjusted its value by calculating the Cohen’s kappa value of a series of classification analyses using different values of the parameter and by choosing the value that had the maximum Cohen kappa value.
2.3.5. Tuning of Parameters and Performance Assessment of the Classification Models
We carried out a stratified 10-fold cross-validation using data corresponding to the classes “Andes mountain forests” and “shrubland,” dividing the data into 10 sub-datasets of approximately the same size and maintaining the ratio of data of each class. Each model was trained using nine folds of the data and validated with the remaining fold.
In order to avoid the overfitting problem, we followed a simple recommended approach to compare classifiers using cross-validation [
41] by carrying out the classifier parameters’ tuning task using only the training data. The procedure is as follows:
We created a training set T = A − k for each of the k subsets of the dataset A
We divided the training set T into subsets t1 and t2; these subsets were used for training and tuning respectively. The subset of variables or features used to fine tune the classifiers are the same set of variables selected for each model, as shown in
Section 3.1.
When the parameters of the classifier were tuned for maximum accuracy, we re-ran each of the models with the initial larger training set T. We chose values of the tuning parameters that maximizes the average of Sensitivity and Specificity metrics. This criterion is recommended for conservation studies where omission error is undesirable [
42].
The classification precision indicators were calculated using the fold k as the validation data.
Mean and standard deviation of the precision indicators were calculated for comparison analysis.
As performance indicators, we used Cohen’s kappa value and the area under the curve (AUC) from the receiver operating characteristic (ROC) theory [
43,
44]. Cohen’s kappa values were calculated using a confusion matrix constructed with the results obtained by classifying the verification data. However, because of the imbalance in the proportion of data between the two land cover types, the result was biased toward the larger one of the two. Thus, analyzing the performance of the models with a probability threshold of 0.5 will under-predict the occurrence of the rarest class [
45,
46]. To avoid this effect, we used the ROC curve to determine the corresponding optimum threshold of each classification analysis by selecting the threshold value that maximizes the average of Sensitivity and Specificity. Using the new calculated threshold value for each classification result in the cross-validation procedure, we constructed the corresponding confusion matrices from which Cohen’s kappa values were calculated. The AUC value is threshold independent, which means it gives a value of overall accuracy based on many different probability thresholds. The value of AUC varies from 0.5 to 1.0 (a perfect fit), and we calculated it with the R software package pROC.
We ranked the performances of all of the models obtained with each fold and calculated the mean value of the ranks obtained per model [
47]; a higher rank (1 being the highest rank) would indicate a higher performance indicator value. The mean ranks of the performance indicators were compared as well as the corresponding mean and standard deviation. Also, Friedman tests were carried out to determine if the various models yielded statistically different AUC and Cohen’s kappa values [
48]. The test was conducted using the mean rank of the performances of all of the models. If the
p-value of the test was significant (
p < 0.05), we could reject the null hypothesis that the difference in classification performances between the models is zero. Then, using the Nemenyi post-hoc test, we carried out a pairwise multiple comparison of ranks and determined which models show a significant performance difference compared to the others by using the critical distance (CD) [
49]. This test is conservative and robust when analyzing a small amount of unbalanced data. The CD value was calculated as follows:
where
N is the number of folds,
k is the number of models to be compared,
α is the confidence level, and
qα is the critical value based on the Studentized range statistic that depends on the significance level
α and
k.
Finally, we attempted to determine if, given the same set of variables, a particular MLA would produce a better classifier for “Andes mountain forest” vs. “shrubs” classification. For this, we conducted Nemenyi post-hoc statistical tests to test to determine if there is a significant difference between the performance indicator values of a particular model produced by RF, SVM, and kNN.