Microarray Analysis of Differentially-Expressed Genes Encoding CYP450 and Phase II Drug Metabolizing Enzymes in Psoriasis and Melanoma
Abstract
:1. Introduction
2. Experimental Section
2.1. Methods
2.2. Validation of Methods
3. Results and Discussion
3.1. Increased Turnover of Vitamin D in Psoriasis
3.2. Differential Regulation of Vitamin A metabolism in Psoriasis versus Melanoma
3.3. Genes Regulating Epidermal Barrier Function Are Up-Regulated in Psoriasis
3.4. Differential Regulation of Antioxidant Potential in Psoriasis versus Melanoma
3.5. Differential Regulation of Cholesterol Catabolism in Psoriasis versus Melanoma
3.6. Regulation of Metabolism of Arachidonic Acid and Xenobiotics in Psoriasis and Melanoma
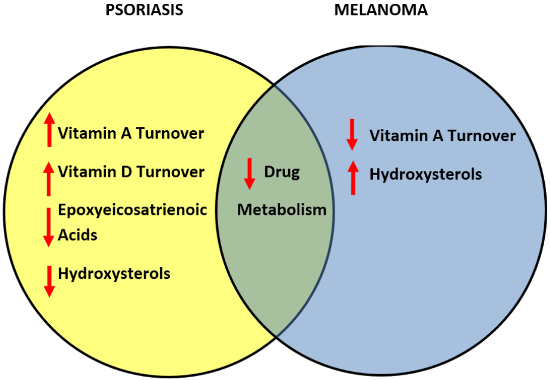
3.7. Comparative Metabolism of Vitamins and Lipids in Psoriasis and Melanomas
3.8. Differentially-Expressed Drug Metabolizing Genes in Psoriasis
3.9. Differentially-Expressed Drug Metabolizing Genes in Melanoma
3.10. Downregulated Drug Metabolizing Genes in Psoriasis and Melanomas
3.11. Drug Development for Psoriasis and Melanomas
4. Conclusions
- Genes controlling abnormal keratinocyte differentiation (CYP24A1) and epidermal barrier function (CYP4F22, SULT2B1) were up-regulated in psoriasis. Therefore, new drugs targeting these genes/enzymes could potentially improve drug penetration, and reduce inflammation and proliferation of keratinocytes and TH-1 cells in psoriatic skin.
- Psoriasis samples showed upregulation of CYP7B1 which could result in increased breakdown of pro-inflammatory hydroxysterols. Melanomas showed the opposite trend. Thus, downregulation of CYP39A1 can lead to accumulation of pro-inflammatory hydroxysterols and enhance melanoma progression.
- This relationship between CYP39A1 expression and inflammation is a novel link between two new hallmarks of cancer (abnormal lipid metabolism and inflammation).
- Psoriasis samples have greater ability to metabolize drugs than melanoma samples. This is because the major DME (CYP3A4) was only down-regulated in melanoma samples.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nebert, D.W.; Wikvall, K.; Miller, W.L. Human cytochromes P450 in health and disease. Philos. Trans. R. Soc. Lond B 2013, 368, 20120431. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Mukhtar, H. Cytochrome P450: A Target for drug development for skin diseases. J. Investig. Dermatol. 2004, 123, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Renton, K.W. Cytochrome P450 regulation and drug biotransformation during inflammation and infection. Curr. Drug Metab. 2004, 5, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Rung, J.; Brazma, A. Reuse of public genome-wide gene expression data. Nat. Rev. Genet. 2012, 14, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Fariñas, M.; Li, K.; Fuentes-Duculan, J.; Hayden, K.; Brodmerkel, C.; Krueger, J.G. Expanding the psoriasis disease profile: Interrogation of the skin and serum of patients with moderate-to-serve psoriasis. J.Investig.Dermatol. 2012, 132, 2552–2564. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Richman, L.; Morehouse, C.; de los Reyes, M. Type I interferon: Potential therapeutic target for psoriasis. PLoS ONE 2008, 3, e2737. [Google Scholar] [CrossRef] [PubMed]
- Bigler, J.; Rand, H.A.; Kerkof, K.; Timour, M.; Russell, C.B. Cross-study homogeneity of psoriasis gene expression in skin across a large expression range. PLoS ONE 2013, 8, e5224. [Google Scholar]
- Raskin, L.; Fullen, D.R.; Giordano, T.J.; Thomas, D.G. Transcriptome profiling identifies HMAG2 as a biomarker of melanoma progression and prognosis. J. Investig. Dermatol. 2013, 133, 2585–2592. [Google Scholar] [CrossRef] [PubMed]
- Kabbarah, O.; Nogueira, C.; Feng, B.; Nazarian, R.M. Integrative genome comparison of primary and metastatic melanomas. PLoS ONE 2010, 5, e10770. [Google Scholar] [CrossRef] [PubMed]
- Talantov, D.; Mazumder, A.; Yu, J.X.; Briggs, T. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin. Cancer Res. 2005, 11, 7234–7242. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Lam, A.; Li, M.C.; Ngan, M.; Menenzes, S.; Zhao, Y. BRB analysis of gene expression data using BRB-Array Tools. Cancer Inform. 2007, 4, 11–17. [Google Scholar]
- Tian, S.; Krueger, J.G.; Li, K.; Jabbari, A.; Brodmerkel, C. Meta-analysis derived (MAD) transcriptome of psoriasis defines the ‘‘Core’’ pathogenesis of disease. PLoS ONE 2012, 7, e44274. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B. The vitamin D3 pathway in human skin and its role for regulation of biological processes. Photochem. Photobiol. 2005, 81, 1246–1251. [Google Scholar] [CrossRef] [PubMed]
- Rashmi, R.; Rao, K.S.; Basavaraj, K.H. A comprehensive review of biomarkers in psoriasis. Clin. Exp. Dermatol. 2009, 34, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Gallego, O.; Ruiz, F.X.; Ardèvol, A.; Domínguez, M.; Alvarez, R.; de Lera, A.R.; Rovira, C.; Farrés, J.; Fita, I.; Parés, X. Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10. Proc. Natl. Acad. Sci. USA 2007, 104, 20764–20769. [Google Scholar] [CrossRef] [PubMed]
- Park, A.L.; Lin, H.K.; Yang, Q.; Sing, C.W.; Fan, M.; Mapstone, T.B.; Gross, N.L.; Gumerlock, M.K.; Martin, M.D.; Rabb, C.H.; et al. Differential expression of type 2 3α/type 5 17β-hydroxysteroid dehydrogenase (AKR1C3) in tumors of the central nervous system. Int. J. Clin. Exp. Pathol. 2010, 3, 743–754. [Google Scholar] [PubMed]
- Mrass, P.; Rendl, M.; Mildner, M.; Gruber, F.; Lengauer, B.; Ballaun, C.; Eckhart, L.; Tschachler, E. Retinoic acid increases the expression of p53 and proapoptotic caspases and sensitizes keratinocytes to apoptosis: A possible explanation for tumor preventive action of retinoids. Cancer Res. 2004, 64, 6542–6548. [Google Scholar] [CrossRef] [PubMed]
- Kelly, E.J.; Nakano, M.; Rohatgil, P.; Yarov-Yarovoy, V.; Rettie, A.E. Finding homes for orphan cytochrome P450s: CYP4V2 and CYP4F22 in disease states. Mol. Invent. 2011, 11, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Garcia, A.; Thomas, C.P.; Keeney, D.S.; Zheng, Y.; Brash, A.R. The importance of the lipoxygenase-hepoxilin pathway in the mammalian epidermal barrier. Biochim. Biophys. Acta. 2014, 1841, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Elias, P.M.; Williams, M.L.; Choi, E.H.; Feingold, K.R. Role of cholesterol sulfate in epidermal structure and function: Lessons from X-linked ichthyosis. Biochim. Biophys. Acta. 2014, 1841, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xu, Y.; Guo, F.; Ning, Y.; Zhi, X.; Yin, L.; Li, X. Hydroxysteroid sulfotransferase SULT2B1b promotes hepatocellular carcinoma cells proliferation in vitro and in vivo . PLoS ONE 2013, 8, e60853. [Google Scholar] [CrossRef] [PubMed]
- Muzio, G.; Maggiora, M.; Paiuzzi, E.; Oraldi, M.; Canuto, R.A. Aldehyde dehydrogenases and cell proliferation. Free Radic. Biol. Med. 2012, 52, 735–746. [Google Scholar] [CrossRef]
- Kitamura, T.; Takagi, S.; Naganuma, T.; Kihara, A. Mouse aldehyde dehydrogenase ALDH3B2 is localized to lipid droplets via two C-terminal tryptophan residues and lipid modification. Biochem. J. 2015, 465, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Langhi, C.; Pedraz-Cuesta, E.; Haro, D.; Marrero, P.F.; Rodríguez, J.C. Regulation of human class I alcohol dehydrogenases by bile acids. J. Lipid Res. 2013, 54, 2475–2484. [Google Scholar] [CrossRef] [Green Version]
- Poli, G.; Biasi, F.; Leonarduzzi, G. Oxysterols in the pathogenesis of major chronic disease. Redox Biol. 2013, 31, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Rosklint, T.; Ohlsson, B.G.; Wiklund, O.; Norén, K.; Hultén, L.M. Oxysterols induce interleukin-1beta production in human macrophages. Eur. J. Clin. Investig. 2002, 32, 35–42. [Google Scholar] [CrossRef]
- Bauman, D.R.; Bitmansour, A.D.; McDonald, J.G.; Thompson, B.M.; Liang, G.; Russell, D.W. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc. Natl. Acad. Sci. USA 2009, 106, 16764–16769. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.T.; Zmijewski, M.A.; Semak, I.; Zbytek, B.; Pisarchik, A.; Li, W.; Zjawiony, J.; Tuckey, R.C. Cytochromes p450 and skin cancer: Role of local endocrine pathways. Anticancer Agents Med. Chem. 2014, 14, 77–96. [Google Scholar] [CrossRef]
- Hennebert, O.; Chalbot, S.; Alran, S.; Morfin, R. Dehydroepiandrosterone 7 alpha-hydroxylation in human tissues: Possible interference with type 1 11beta-hydroxysteroid dehydrogenase-mediated processes. J. Steroid Biochem. Mol. Biol. 2007, 104, 326–333. [Google Scholar] [CrossRef]
- Khenjanta, C.; Thanan, R.; Jusakul, A.; Techasen, A.; Jamnongkan, W.; Namwat, N.; Loilome, W.; Pairojkul, C.; Yongvanit, P. Association of CYP39A1, RUNX2 and oxidized alpha-1 antitrypsin expression in relation to cholangiocarcinoma progression. Asian Pac. J. Cancer Prev. 2014, 23, 10187–10192. [Google Scholar] [CrossRef]
- Nithipatikom, K.; Moore, J.M.; Isbell, M.A.; Falck, J.R.; Gross, G.J. Epoxyeicosatrienoic acids in cardioprotection: Ischemic versus reperfusion injury. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Ladd, P.A.; Du, L.; Capdevila, J.H.; Mernaugh, R.; Keeney, D.S. Epoxyeicosatrienoic acids activate transglutaminases in situ and induce cornification of epidermal keratinocytes. J. Biol. Chem. 2003, 278, 35184–35192. [Google Scholar] [CrossRef] [PubMed]
- Baer, B.R.; Rettie, A.E. CYP4B1: An enigmatic P450 at the interface between xenobiotic and endobiotic metabolism. Drug Metab. Rev. 2006, 38, 451–476. [Google Scholar] [CrossRef] [PubMed]
- Wiek, C.; Schmidt, E.M.; Roellecke, K.; Freund, M.; Nakano, M.; Kelly, E.J.; Kaisers, W.; Yarov-Yarovoy, V.; Kramm, C.M.; Rettie, A.E.; et al. Identification of amino acid determinants in CYP4B1 for optimal catalytic processing of 4-ipomeanol. Biochem. J. 2015, 465, 103–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldstein, J.A. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br. J. Clin. Pharmacol. 2001, 52, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Marill, J.; Cresteil, T.; Lanotte, M.; Chabot, G.G. Identification of humancytochromeP450s involved in the formation of all-trans-retinoicacid principal metabolites. Mol. Pharmacol. 2000, 58, 1341–1348. [Google Scholar] [PubMed]
- Martínez-Jiménez, C.P.; Jover, R.; Donato, M.T.; Castell, J.V.; Gómez-Lechón, M.J. Transcriptional regulation and expression of CYP3A4 in hepatocytes. Curr. Drug Metab. 2007, 8, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, P.F.; Chun, Y.J.; Kim, D.; Gillam, E.M.; Shimada, T. Cytochrome P450 1B1: A target for inhibition in anticarcinogenesis strategies. Mutat. Res. 2003, 523–524, 173–182. [Google Scholar] [CrossRef]
- Shimada, T.; Fujii-Kuriyama, Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004, 95, 1–6. [Google Scholar] [CrossRef]
- Katiyar, S.K.; Matsui, M.S.; Mukhtar, H. Ultraviolet-B exposure of human skin induces cytochromes P450 1A1 and 1B1. J. Investig. Dermatol. 2000, 114, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Buters, J.T.M.; Sakai, S.; Richter, T.; Pineau, T.; Alexander, D.L.; Savas, U.; Doehmer, J.; Ward, J.M.; Jefcoate, C.R.; Gonzalez, F.Z. Cytochrome P450 CYP1B1 determines susceptibility to 7,12-dimethylbenz[a]anthracene-induced lymphomas. Proc. Natl. Acad. Sci. USA 1999, 96, 1977–1982. [Google Scholar] [CrossRef] [PubMed]
- Wachsman, W.; Morhenn, V.; Palmer, T.; Walls, L.; Hata, T.; Zalla, J.; Scheinberg, R.; Sofen, H.; Mraz, S.; Gross, K.; et al. Noninvasive genomic detection of melanoma. Br. J. Dermatol. 2011, 164, 797–806. [Google Scholar] [CrossRef]
- Saini, S.; Hirata, H.; Majid, S.; Dahiya, R. Functional significance of cytochrome P450 1B1 in endometrial carcinogenesis. Cancer Res. 2009, 69, 7038–7045. [Google Scholar] [CrossRef] [PubMed]
- McFadyen, M.C.; McLeod, H.L.; Jackson, F.C.; Melvin, W.T.; Doehmer, J.; Murray, G.I. Cytochrome P450 CYP1B1 protein expression: A novel mechanism of anticancer drug resistance. Biochem. Pharmacol. 2001, 62, 207–212. [Google Scholar] [CrossRef]
- Kuehl, P.; Zhang, J.; Lin, Y.; Lamba, J.; Assem, M.; Schuetz, J.; Watkins, P.B.; Daly, A.; Wrighton, S.A.; Hall, S.D.; et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 2001, 27, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Hustert, E.; Haberl, M.; Burk, O.; Wolbold, R.; He, Y.-Q.; Klein, K.; Nuessler, A.C.; Neuhaus, P.; Klattig, J.; Eiselt, R.; et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 2001, 11, 773–779. [Google Scholar] [CrossRef] [PubMed]
- van Eijl, S.; Zhu, Z.; Cupitt, J.; Gierula, M.; Götz, C.; Fritsche, E.; Edwards, R.J. Elucidation of xenobiotic metabolism pathways in human skin and human skin models by proteomic profiling. PLoS ONE 2012, 7, e41721. [Google Scholar] [CrossRef] [PubMed]
- Brennan, B.J.; Xu, Z.X.; Grippo, J.F. Effect of peginterferon alfa-2a (40KD) on cytochrome P450 isoenzyme activity. Br. J. Clin. Pharmacol. 2013, 75, 497–506. [Google Scholar] [CrossRef]
- Kimm, S.; Östör, A.J.; Nisar, M.K. Interleukin-6 and cytochrome-P450, reason for concern? Rheumatol. Int. 2012, 32, 2601–2604. [Google Scholar] [CrossRef] [PubMed]
- Brănişteanu, D.E.; Voicu, C.M.; Creţu, A.; Dimitriu, A.; Luca, M.C.; Sălăvăstru, C.M. Adverse reactions of biological therapy for psoriasis. Rev. Med. Chir. Soc. Med. Nat. Iasi. 2015, 119, 38–44. [Google Scholar] [PubMed]
- Harvey, R.D.; Morgan, E.T. Cancer, inflammation, and therapy: Effects on cytochrome p450-mediated drug metabolism and implications for novel immunotherapeutic agents. Clin. Pharmacol. Ther. 2014, 96, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.T.; Lathia, C.; Frye, R.F.; Schuchter, L.; Redlinger, M.; Rosen, M.; O'Dwyer, P.J. Interaction of sorafenib and cytochrome P450 isoenzymes in patients with advanced melanoma: A phase I/II pharmacokinetic interaction study. Cancer Chemother. Pharmacol. 2011, 68, 1111–1118. [Google Scholar] [CrossRef] [PubMed]


| Psoriasis | GSE 14905 | GSE 30999 | GSE 41662 | GSE 34248 |
|---|---|---|---|---|
| No. Arrays | 21 (NS) 33 (PS) | 85(NS) 85(PS) | 24(NS) 24(PS) | 14(NS) 14(PS) |
| No. Input Genes | 4994 | 8961 | 12416 | 9596 |
| No. DEG | 3328 | 6826 | 4227 | 2186 |
| Melanoma | GSE15605 | GDS1375 | GSE46517 | |
| No. Arrays | 16 (NS) 46 (PM) | 7 (NS) 31 (PM) | 7 (NS) 45 (PM) | |
| No. Input Genes | 11938 | 6514 | 2378 | |
| No. DEG | 4279 | 3282 | 772 |
| Gene Symbol | Probe Set ID | Mean ± S.D Fold Change | Mean Parametric p-Value | Function of Enzyme Encoded by Gene | Impact of Altered Gene Expression On Disease |
|---|---|---|---|---|---|
| CYP27B1 | 205676_at | 3.13 ± 0.43 | 3.20 × 10−4 | Catalyzes activation of vitamin D | Vitamin D promotes normal keratinocyte differentiation |
| CYP24A1 | 206504_at | 8.80 ± 0.68 | 9.00 × 10−6 | Catabolizes active vitamin D | Depleted vitamin D leads to abnormal keratinocyte differentiation |
| CYP4F22 | 244692_at | 5.00 ± 1.25 | <1 × 10−7 | Regulates hepoxillins in permeability barrier | Impaired corneocyte structure and barrier function |
| CYP7B1 | 207386_at | 2.60 ± 0.80 | 9.02 × 10−5 | Inactivates pro-inflammatory 25 and 27-hydroxysterols | Potential Anti-inflammatory effect. Interferes with action of Glucocorticoid Drugs |
| CYP2C18 | 208126_s_at | 4.02 ± 0.89 | 2.32 × 10−4 | Metabolizes important drugs and vitamin A | Vitamin A metabolites can inhibit keratinocyte proliferation |
| AKR1B10 | 206561_s_at | 23.8 ± 15.80 | <1 × 10−7 | Trans-retinaldehyde-reductase degrades vitamin A | Decreased vitamin A can stimulate keratinocyte proliferation |
| SULT2B1 | 205759_s_at | 3.31 ± 0.46 | <1 × 10−7 | Sulfo-transferase sulfates cholesterol | Excess cholesterol sulfate promotes abnormal barrier |
| CYP2J2 | 205073_at | 0.37 ± 0.065 | 3.96 × 10−4 | Produces EETs with anti-inflammatory effects. | Downregulation decreases EETs . Insufficient cornification and increased inflammation can occur |
| CYP4B1 | 210096_at | 0.39 ± 0.04 | 1.26 × 10−4 | Synthesizes eicosanoids. Activates specific xenobiotics | Downregulation has no impact since human CYP4B1 protein lacks function |
| CYP1B1 | 202436_s_at | 0.37 ± 0.18 | 3.26 × 10−4 | Activates specific xenobiotics and mutagens | Downregulation can protect against toxins and carcinogens |
| CYP3A5 | 205765_at | 0.56 ± 0.05 | 9.73 × 10−4 | Metabolizes important drugs | Downregulation can decrease drug metabolism |
| Gene Symbol | Probe Set ID | Mean ± SD Fold Change | Mean Parametric p-Value | Function of Enzyme Encoded by Gene | Impact of Altered Gene Expression On Disease |
|---|---|---|---|---|---|
| CYP4B1 | 210096_at | 0.09 ± 0.05 | <1 × 10−7 | Synthesizes inflammatory eicosanoids. Activates specific xenobiotics | Downregulation has no impact since human CYP4B1 protein lacks function |
| CYP39A1 | 220432_s_at | 0.27 ± 0.15 | 2.08 × 10−4 | Inactivates pro-inflammatory 24-hydroxysterols | Downregulation can have pro-Inflammatory effect and increase metastasis |
| CYP1B1 | 202437_s_at | 0.24 ± 0.11 | 1.56 × 10−5 | Activates specific xenobiotics and mutagens | Downregulation can protect against toxins and carcinogens |
| CYP3A4 | 205999_x_at | 0.31 ± 0.04 | 1.50 × 10−5 | Metabolizes 50% of important drugs | Downregulation can significantly decrease drug metabolizing capacity |
| CYP3A5 | 214234_s_at | 0.19 ± 0.15 | 3.33 × 10−5 | Functionally similar to CYP3A4 | Downregulation can decrease drug metabolism |
| AKR1C3 | 209160_at | 0.19 ± 0.16 | 1.80 × 10−5 | Trans-retinaldehyde-reductase which degrades vitamin A | Downregulation can stabilize vitamin A levels and limit cell proliferation |
| SULT2B1 | 205759_s_at | 0.30 ± 0.14 | 5.95 × 10−5 | Sulfo-transferase which sulfates cholesterol | Downregulation can lead to decreased proliferation |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sumantran, V.N.; Mishra, P.; Bera, R.; Sudhakar, N. Microarray Analysis of Differentially-Expressed Genes Encoding CYP450 and Phase II Drug Metabolizing Enzymes in Psoriasis and Melanoma. Pharmaceutics 2016, 8, 4. https://doi.org/10.3390/pharmaceutics8010004
Sumantran VN, Mishra P, Bera R, Sudhakar N. Microarray Analysis of Differentially-Expressed Genes Encoding CYP450 and Phase II Drug Metabolizing Enzymes in Psoriasis and Melanoma. Pharmaceutics. 2016; 8(1):4. https://doi.org/10.3390/pharmaceutics8010004
Chicago/Turabian StyleSumantran, Venil N., Pratik Mishra, Rakesh Bera, and Natarajan Sudhakar. 2016. "Microarray Analysis of Differentially-Expressed Genes Encoding CYP450 and Phase II Drug Metabolizing Enzymes in Psoriasis and Melanoma" Pharmaceutics 8, no. 1: 4. https://doi.org/10.3390/pharmaceutics8010004