Novel Multi-Responsive Hyperbranched Polyelectrolyte Polyplexes as Potential Gene Delivery Vectors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Hyperbranched P(OEGMA-co-DIPAEMA) Polyelectrolyte Copolymers Synthesis
2.3. Preparation of P(OEGMA-co-DIPAEMA)/DNA Polyplexes in Aqueous Media
2.4. FBS Interactions with P(OEGMA-co-DIPAEMA)/DNA Polyplexes
2.5. MTT Assay
2.6. Methods
2.6.1. Dynamic Light Scattering
2.6.2. Electrophoretic Light Scattering
2.6.3. Fluorescence Spectroscopy
3. Results and Discussion
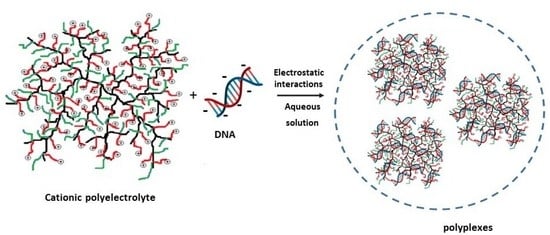
3.1. Complexation Behaviour of Hyperbranched Polyelectrolyte Copolymers with DNA in Aqueous Solutions
3.2. Effect of Solution Ionic Strength on the Formed Polyplexes
3.3. Temporal Stability Studies of the Polyplexes
3.4. Ethidium Bromide Quenching Assay on Polyplexes
3.5. FBS Interactions with Polyplexes
3.6. In Vitro Cytotoxicity Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mintzer, M.A.; Simanek, E.E. Synthetic vectors for gene delivery. Chem. Rev. 2009, 109, 259–302. [Google Scholar] [CrossRef] [PubMed]
- Pack, D.W.; Hoffman, A.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Silva, M.; Jarak, I.; Alvarez-Lorenzo, C.; Concheiro, A.; Santos, A.C.; Veiga, F.; Figueiras, A. Micelleplexes as nucleic acid delivery systems for cancer-targeted therapies. J. Control Release 2020, 323, 442–462. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, C.; Liang, W. Development of RNAi technology for targeted therapy--a track of siRNA based agents to RNAi therapeutics. J. Control Release 2014, 193, 270–281. [Google Scholar] [CrossRef]
- Nishiyama, N.; Matsumura, Y.; Kataoka, K. Development of polymeric micelles for targeting intractable cancers. Cancer Sci. 2016, 107, 867–874. [Google Scholar] [CrossRef] [Green Version]
- Cabral, H.; Miyata, K.; Osada, K.; Kataoka, K. Block Copolymer Micelles in Nanomedicine Applications. Chem. Rev. 2018, 118, 6844–6892. [Google Scholar] [CrossRef] [Green Version]
- Tros de Ilarduya, C.; Sun, Y.; Düzgüneş, N. Gene delivery by lipoplexes and polyplexes. Eur. J. Pharm. Sci. 2010, 40, 159–170. [Google Scholar] [CrossRef]
- Zhang, X.X.; McIntosh, T.J.; Grinstaff, M.W. Functional lipids and lipoplexes for improved gene delivery. Biochimie 2012, 94, 42–58. [Google Scholar] [CrossRef] [Green Version]
- Selianitis, D.; Kafetzi, M.; Pippa, N.; Pispas, S.; Gazouli, M. Lipoplexes and polyplexes for targeted gene delivery. In Nanotechnology in the Life Sciences, 1st ed.; Barabadi, H., Mostafavi, E., Saravanan, M., Eds.; Springer: Cham, Switzerland, 2022; pp. 65–92. [Google Scholar]
- Triantafyllopoulou, E.; Pippa, N.; Demetzos, C. Protein-liposome interactions: The impact of surface charge and fluidisation effect on protein binding. J. Liposome Res. 2023, 33, 77–88. [Google Scholar] [CrossRef]
- Zu, H.; Gao, D. Non-viral Vectors in Gene Therapy: Recent Development, Challenges, and Prospects. AAPS J. 2021, 23, 78. [Google Scholar] [CrossRef] [PubMed]
- Rezaee, M.; Oskuee, R.K.; Nassirli, H.; Malaekeh-Nikouei, B. Progress in the development of lipopolyplexes as efficient non-viral gene delivery systems. J. Control Release 2016, 236, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo-Rodríguez, A.; Solinís, M.Á.; Rodríguez-Gascón, A. Applications of lipid nanoparticles in gene therapy. Eur. J. Pharm. Biopharm. 2016, 109, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Haladjova, E.; Panseri, S.; Montesi, M.; Rossi, A.; Skandalis, A.; Pispas, S.; Rangelov, S. Influence of DNA Type on the Physicochemical and Biological Properties of Polyplexes Based on Star Polymers Bearing Different Amino Functionalities. Polymers 2023, 15, 894. [Google Scholar] [CrossRef]
- Dinari, A.; Moghadam, T.T.; Abdollahi, M.; Sadeghizadeh, M. Synthesis and Characterization of a Nano-Polyplex system of GNRs-PDMAEA-pDNA: An Inert Self-Catalyzed Degradable Carrier for Facile Gene Delivery. Sci. Rep. 2018, 8, 8112. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Jung, S.; Si, G.; Cheng, R.; Meng, F.; Zhu, X.; Park, T.G.; Zhong, Z. Cationic methacrylate copolymers containing primary and tertiary amino side groups: Controlled synthesis via RAFT polymerization, DNA condensation, and in vitro gene transfection. J. Polym. Sci. A Polym. Chem. 2010, 48, 2869–2877. [Google Scholar] [CrossRef]
- Rosselgong, J.; Williams, E.G.; Le, T.P.; Grusche, F.; Hinton, T.M.; Tizard, M.; Gunatillake, P.; Thang, S.H. Core degradable star RAFT polymers: Synthesis, polymerization, and degradation studies. Macromolecules 2013, 46, 9181–9188. [Google Scholar] [CrossRef]
- Alfurhood, J.A.; Bachler, P.R.; Sumerlin, B.S. Hyperbranched polymers via RAFT self-condensing vinyl polymerization. Polym. Chem. 2016, 7, 3361–3369. [Google Scholar] [CrossRef]
- Kondinskaia, D.A.; Gurtovenko, A.A. Supramolecular complexes of DNA with cationic polymers: The effect of polymer concentration. Polymer 2018, 142, 277–284. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, N. Cationic polymer optimization for efficient gene delivery. Mini Rev. Med. Chem. 2010, 10, 108–125. [Google Scholar] [CrossRef]
- Garamus, V.M.; Maksimova, T.V.; Kautz, H.; Barriau, E.; Frey, H.; Schlotterbeck, U.; Mecking, S.; Richtering, W. Hyperbranched polymers: Structure of hyperbranched polyglycerol and amphiphilic poly (glycerol ester) s in dilute aqueous and nonaqueous solution. Macromolecules 2004, 37, 8394–8399. [Google Scholar] [CrossRef] [Green Version]
- Wilms, D.; Stiriba, S.-E.; Frey, H. Hyperbranched polyglycerols: From the controlled synthesis of biocompatible polyether polyols to multipurpose applications. Acc. Chem. Res. 2010, 43, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Dalakoglou, G.; Karatasos, K.; Lyulin, S.; Larin, S.; Darinskii, A.; Lyulin, A. Conformational effects in non-stoichiometric complexes of two hyperbranched molecules with a linear polyelectrolyte. Polymers 2012, 4, 240–255. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Zhu, J.; Song, X.; Wen, Y.; Li, J. Smart Hydrogel Formed by Alginate-g-Poly (N-isopropylacrylamide) and Chitosan through Polyelectrolyte Complexation and Its Controlled Release Properties. Gels 2022, 8, 441. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, W.; Liu, J.; Zhu, X.; Yan, D. Self-assembly of hyperbranched polymers and its biomedical applications. Adv. Mater. 2010, 22, 4567–4590. [Google Scholar] [CrossRef]
- Liu, J.; Huang, W.; Pang, Y.; Yan, D. Hyperbranched polyphosphates: Synthesis, functionalization and biomedical applications. Chem. Soc. Rev. 2015, 44, 3942–3953. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.; Scholz, C. Highly branched polymers based on poly (amino acid) s for biomedical application. Nanomaterials 2021, 11, 1119. [Google Scholar] [CrossRef]
- Segawa, Y.; Higashihara, T.; Ueda, M. Synthesis of hyperbranched polymers with controlled structure. Polym. Chem. 2013, 4, 1746–1759. [Google Scholar] [CrossRef]
- Zhang, D.; Jia, D.; Zhou, Z. Synthesis and characterization of low viscosity aromatic hyperbranched polyester epoxy resin. Macromol. Res. 2009, 17, 289–295. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Qiu, S.; Yu, B.; Wang, J.; Wang, C.; Zeng, W.; Hu, Y. Economical and environment-friendly synthesis of a novel hyperbranched poly (aminomethylphosphine oxide-amine) as co-curing agent for simultaneous improvement of fire safety, glass transition temperature and toughness of epoxy resins. Chem. Eng. J. 2017, 322, 618–631. [Google Scholar] [CrossRef]
- Irfan, M.; Seiler, M. Encapsulation using hyperbranched polymers: From research and technologies to emerging applications. Ind. Eng. Chem. Res. 2010, 49, 1169–1196. [Google Scholar] [CrossRef]
- Seleci, M.; Seleci, D.A.; Ciftci, M.; Odaci Demirkol, D.; Stahl, F.; Timur, S.; Scheper, D.; Yagci, Y. Nanostructured amphiphilic star-hyperbranched block copolymers for drug delivery. Langmuir 2015, 31, 4542–4551. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, H.; Su, Y.; Qiu, F.; Zhu, L.; Huan, X.; Guo, F.; Zhu, X. Supramolecular amphiphilic multiarm hyperbranched copolymer: Synthesis, self-assembly and drug delivery applications. Polym. Chem. 2013, 4, 85–94. [Google Scholar] [CrossRef]
- Selianitis, D.; Forys, A.; Trzebicka, B.; Alemayehu, A.; Tyrpekl, V.; Pispas, S. Amphiphilic P (OEGMA-Co-DIPAEMA) Hyperbranched Copolymer/Magnetic Nanoparticle Hybrid Nanostructures by Co-Assembly. Nanomanufacturing 2022, 2, 53–68. [Google Scholar] [CrossRef]
- Ahmed, M.; Narain, R. The effect of molecular weight, compositions and lectin type on the properties of hyperbranched glycopolymers as non-viral gene delivery systems. Biomaterials 2012, 33, 3990–4001. [Google Scholar] [CrossRef]
- Cook, A.B.; Peltier, R.; Zhang, J.; Gurnani, P.; Tanaka, J.; Burns, J.; Hartlieb, M.; Perrier, S. Hyperbranched poly (ethylenimine-co-oxazoline) by thiol–yne chemistry for non-viral gene delivery: Investigating the role of polymer architecture. Polym. Chem. 2019, 10, 1202–1212. [Google Scholar] [CrossRef]
- Rodrigues, P.R.; Wang, X.; Li, Z.; Lyu, J.; Wang, W.; Vieira, R.P. A new nano hyperbranched β-pinene polymer: Controlled synthesis and nonviral gene delivery. Colloids Surf. B. 2023, 222, 113032. [Google Scholar] [CrossRef] [PubMed]
- Hatamzadeh, M.; Sarvari, R.; Massoumi, B.; Agbolaghi, S.; Samadian, F. Liver tissue engineering via hyperbranched polypyrrole scaffolds. Int. J. Polym. Mater. Polym. Biomater. 2020, 69, 1112–1122. [Google Scholar] [CrossRef]
- Guo, B.; Ma, P.X. Synthetic biodegradable functional polymers for tissue engineering: A brief review. Sci. China Chem. 2014, 57, 490–500. [Google Scholar] [CrossRef]
- Saadati, A.; Hasanzadeh, M.; Seidi, F. Biomedical application of hyperbranched polymers: Recent Advances and challenges. TrAC Trends Anal. Chem. 2021, 142, 116308. [Google Scholar] [CrossRef]
- Banik, B.L.; Fattahi, P.; Brown, J.L. Polymeric nanoparticles: The future of nanomedicine. WIREs Nanomed. Nanobiotechnol. 2016, 8, 271–299. [Google Scholar] [CrossRef]
- Das, S.S.; Bharadwaj, P.; Bilal, M.; Barani, M.; Rahdar, A.; Taboada, P.; Bungau, S.; Kyzas, G.Z. Stimuli-responsive polymeric nanocarriers for drug delivery, imaging, and theragnosis. Polymers 2020, 12, 1397. [Google Scholar] [CrossRef] [PubMed]
- Pasparakis, G.; Tsitsilianis, C. LCST polymers: Thermoresponsive nanostructured assemblies towards bioapplications. Polymer 2020, 211, 123146. [Google Scholar] [CrossRef]
- Selianitis, D.; Pispas, S. PDEGMA-b-PDIPAEMA copolymers via RAFT polymerization and their pH and thermoresponsive schizophrenic self-assembly in aqueous media. J. Polym. Sci. 2020, 58, 1867–1880. [Google Scholar] [CrossRef]
- Tomara, M.; Selianitis, D.; Pispas, S. Dual-Responsive Amphiphilic P (DMAEMA-co-LMA-co-OEGMA) Terpolymer Nano-Assemblies in Aqueous Media. Nanomaterials 2022, 12, 3791. [Google Scholar] [CrossRef]
- Selianitis, D.; Pispas, S. Multi-responsive poly (oligo (ethylene glycol) methyl methacrylate)-co-poly (2-(diisopropylamino) ethyl methacrylate) hyperbranched copolymers via reversible addition fragmentation chain transfer polymerization. Polym. Chem. 2021, 12, 6582–6593. [Google Scholar] [CrossRef]
- Thavanesan, T.; Herbert, C.; Plamper, F.A. Insight in the phase separation peculiarities of poly (dialkylaminoethyl methacrylate) s. Langmuir 2014, 30, 5609–5619. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; Yu, Y.; Zhang, W. Thermoresponsive Polymers Based on Tertiary Amine Moieties. Macromol. Rapid Commun. 2021, 42, 2100504. [Google Scholar] [CrossRef]
- Hu, Y.Q.; Kim, M.S.; Kim, B.S.; Lee, D.S. Synthesis and pH-dependent micellization of 2-(diisopropylamino) ethyl methacrylate based amphiphilic diblock copolymers via RAFT polymerization. Polymer 2007, 48, 3437–3443. [Google Scholar] [CrossRef]
- Haladjova, E.; Mountrichas, G.; Pispas, S.; Rangelov, S. Poly(vinyl benzyl trimethylammonium chloride) Homo and Block Copolymers Complexation with DNA. J. Phys. Chem. B 2016, 120, 2586–2595. [Google Scholar] [CrossRef]
- Geall, A.J.; Blagbrough, I.S. Rapid and sensitive ethidium bromide fluorescencequenching assay of polyamine conjugate–DNA interactions for the analysis of lipoplex formation in gene therapy. J. Pharm. Biomed. Anal. 2000, 22, 839–859. [Google Scholar] [CrossRef] [PubMed]









| Sample | Mw a (g/mol) (×104) | Mw b (g/mol) (×104) | Mw/Mn b | %wt PDIPAEMA c | %wt POEGMA c |
|---|---|---|---|---|---|
| HBC-1 | 33.3 | 1.2 | 1.21 | 10 | 90 |
| HBC-2 | 48.6 | 0.8 | 1.19 | 29 | 71 |
| HBC-3 | 35.1 | 1.1 | 1.24 | 54 | 46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selianitis, D.; Katifelis, H.; Gazouli, M.; Pispas, S. Novel Multi-Responsive Hyperbranched Polyelectrolyte Polyplexes as Potential Gene Delivery Vectors. Pharmaceutics 2023, 15, 1627. https://doi.org/10.3390/pharmaceutics15061627
Selianitis D, Katifelis H, Gazouli M, Pispas S. Novel Multi-Responsive Hyperbranched Polyelectrolyte Polyplexes as Potential Gene Delivery Vectors. Pharmaceutics. 2023; 15(6):1627. https://doi.org/10.3390/pharmaceutics15061627
Chicago/Turabian StyleSelianitis, Dimitrios, Hector Katifelis, Maria Gazouli, and Stergios Pispas. 2023. "Novel Multi-Responsive Hyperbranched Polyelectrolyte Polyplexes as Potential Gene Delivery Vectors" Pharmaceutics 15, no. 6: 1627. https://doi.org/10.3390/pharmaceutics15061627