Aptamers Enhance Oncolytic Viruses’ Antitumor Efficacy
Abstract
:1. Introduction
2. Oncolytic Viruses
3. Aptamers
4. Applications of Aptamers
4.1. Use of the Aptamers as Drug Delivery Vehicles
4.2. The Use of the Aptamers for Diagnostics and Detection of Tumor Cells
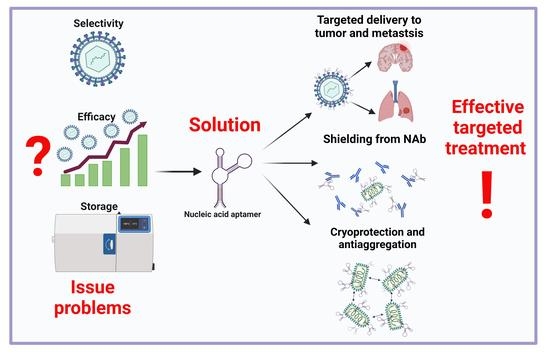
4.3. Aptamers Used to Increase the Antitumor Efficacy of an Oncolytic Virus
4.3.1. Use of the Aptamers to Shield the Oncolytic Virus from the Binding by Neutralizing Antibodies
4.3.2. Aptamers Change the Binding Specificity of Viruses
4.3.3. Aptamers as Cryoprotectors of Oncolytic Viruses and Aggregation Inhibitors
5. Conclusions
- (1)
- Shielding the virus from neutralizing antibodies;
- (2)
- Increasing the targeting and specificity of the viral particles;
- (3)
- Cryoprotective and anti-aggregative properties of the oligonucleotides bound to oncolytic viruses during transportation and storage.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. Available online: https://gco.iarc.fr/today (accessed on 29 September 2021).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.J.; Peng, K.-W.; Bell, J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012, 30, 658–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, G.; He, X.; Sun, Q.; Chen, S.; Wan, K.; Xu, X.; Feng, X.; Li, P.; Chen, B.; Xiong, M. The Oncolytic Virus in Cancer Diagnosis and Treatment. Front. Oncol. 2020, 10, 1786. [Google Scholar] [CrossRef] [PubMed]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Sharon, D.; Kamen, A. Advancements in the design and scalable production of viral gene transfer vectors. Biotechnol. Bioeng. 2018, 115, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, C.; Galanis, E. Interferon signaling predicts response to oncolytic virotherapy. Oncotarget 2019, 10, 1544–1545. [Google Scholar] [CrossRef]
- Lin, W.; Zhao, Y.; Zhong, L. Current strategies of virotherapy in clinical trials for cancer treatment. J. Med. Virol. 2021, 93, 4668–4692. [Google Scholar] [CrossRef]
- Fukuhara, H.; Ino, Y.; Todo, T. Oncolytic virus therapy: A new era of cancer treatment at dawn. Cancer Sci. 2016, 107, 1373–1379. [Google Scholar] [CrossRef]
- Mullen, J.T.; Tanabe, K.K. Viral Oncolysis. Oncologist 2002, 7, 106–119. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Kohlhapp, F.J.; Zloza, A. Oncolytic viruses: A new class of immunotherapy drugs. Nat. Rev. Drug Discov. 2015, 14, 642–662. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Song, K.; Li, S. The Role of Ubiquitination in NF-κB Signaling during Virus Infection. Viruses 2021, 13, 145. [Google Scholar] [CrossRef]
- Cesaro, T.; Michiels, T. Inhibition of PKR by Viruses. Front. Microbiol. 2021, 12, 757238. [Google Scholar] [CrossRef]
- Lemos de Matos, A.; Franco, L.S.; McFadden, G. Oncolytic Viruses and the Immune System: The Dynamic Duo. Mol. Ther.-Methods Clin. Dev. 2020, 17, 349–358. [Google Scholar] [CrossRef] [Green Version]
- Tollefson, A.E.; Scaria, A.; Hermiston, T.W.; Ryerse, J.S.; Wold, L.J.; Wold, W.S. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 1996, 70, 2296–2306. [Google Scholar] [CrossRef] [Green Version]
- Lavoie, J.N.; Nguyen, M.; Marcellus, R.C.; Branton, P.E.; Shore, G.C. E4orf4, a Novel Adenovirus Death Factor That Induces p53-independent Apoptosis by a Pathway That Is Not Inhibited by zVAD-fmk. J. Cell Biol. 1998, 140, 637–645. [Google Scholar] [CrossRef]
- Hu, H.; Zhang, S.; Cai, L.; Duan, H.; Li, Y.; Yang, J.; Wang, Y.; Liu, B.; Dong, S.; Fang, Z.; et al. A novel cocktail therapy based on quintuplet combination of oncolytic herpes simplex virus-2 vectors armed with interleukin-12, interleukin-15, GM-CSF, PD1v, and IL-7 × CCL19 results in enhanced antitumor efficacy. Virol. J. 2022, 19, 74. [Google Scholar] [CrossRef]
- Jhawar, S.R.; Thandoni, A.; Bommareddy, P.K.; Hassan, S.; Kohlhapp, F.J.; Goyal, S.; Schenkel, J.M.; Silk, A.W.; Zloza, A. Oncolytic Viruses—Natural and Genetically Engineered Cancer Immunotherapies. Front. Oncol. 2017, 7, 202. [Google Scholar] [CrossRef] [Green Version]
- Veinalde, R.; Grossardt, C.; Hartmann, L.; Bourgeois-Daigneault, M.-C.; Bell, J.C.; Jäger, D.; von Kalle, C.; Ungerechts, G.; Engeland, C.E. Oncolytic measles virus encoding interleukin-12 mediates potent antitumor effects through T cell activation. Oncoimmunology 2017, 6, e1285992. [Google Scholar] [CrossRef] [Green Version]
- Cervera-Carrascon, V.; Siurala, M.; Santos, J.M.; Havunen, R.; Tähtinen, S.; Karell, P.; Sorsa, S.; Kanerva, A.; Hemminki, A. TNFa and IL-2 armed adenoviruses enable complete responses by anti-PD-1 checkpoint blockade. Oncoimmunology 2018, 7, e1412902. [Google Scholar] [CrossRef]
- Ferrucci, P.F.; Pala, L.; Conforti, F.; Cocorocchio, E. Talimogene Laherparepvec (T-VEC): An Intralesional Cancer Immunotherapy for Advanced Melanoma. Cancers 2021, 13, 1383. [Google Scholar] [CrossRef] [PubMed]
- Rojas, J.M.; Avia, M.; Martín, V.; Sevilla, N. IL-10: A Multifunctional Cytokine in Viral Infections. J. Immunol. Res. 2017, 2017, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillespie, G.Y.; Roth, J.; Friedman, G.; Smith, T. Oncolytic viral therapy: Targeting cancer stem cells. Oncolytic Virotherapy 2014, 3, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaurasiya, S.; Chen, N.; Warner, S. Oncolytic Virotherapy versus Cancer Stem Cells: A Review of Approaches and Mechanisms. Cancers 2018, 10, 124. [Google Scholar] [CrossRef] [Green Version]
- Spiesschaert, B.; Angerer, K.; Park, J.; Wollmann, G. Combining Oncolytic Viruses and Small Molecule Therapeutics: Mutual Benefits. Cancers 2021, 13, 3386. [Google Scholar] [CrossRef]
- Chianese, A.; Santella, B.; Ambrosino, A.; Stelitano, D.; Rinaldi, L.; Galdiero, M.; Zannella, C.; Franci, G. Oncolytic Viruses in Combination Therapeutic Approaches with Epigenetic Modulators: Past, Present, and Future Perspectives. Cancers 2021, 13, 2761. [Google Scholar] [CrossRef]
- Andtbacka, R.H.I.; Kaufman, H.L.; Collichio, F.; Amatruda, T.; Senzer, N.; Chesney, J.; Delman, K.A.; Spitler, L.E.; Puzanov, I.; Agarwala, S.S.; et al. Talimogene Laherparepvec Improves Durable Response Rate in Patients with Advanced Melanoma. J. Clin. Oncol. 2015, 33, 2780–2788. [Google Scholar] [CrossRef]
- Eissa, I.; Bustos-Villalobos, I.; Ichinose, T.; Matsumura, S.; Naoe, Y.; Miyajima, N.; Morimoto, D.; Mukoyama, N.; Zhiwen, W.; Tanaka, M.; et al. The Current Status and Future Prospects of Oncolytic Viruses in Clinical Trials against Melanoma, Glioma, Pancreatic, and Breast Cancers. Cancers 2018, 10, 356. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Fang, H. Clinical Trials with Oncolytic Adenovirus in China. Curr. Cancer Drug Targets 2007, 7, 141–148. [Google Scholar] [CrossRef]
- Doniņa, S.; Strēle, I.; Proboka, G.; Auziņš, J.; Alberts, P.; Jonsson, B.; Venskus, D.; Muceniece, A. Adapted ECHO-7 virus Rigvir immunotherapy (oncolytic virotherapy) prolongs survival in melanoma patients after surgical excision of the tumour in a retrospective study. Melanoma Res. 2015, 25, 421–426. [Google Scholar] [CrossRef] [Green Version]
- Frampton, J.E. Teserpaturev/G47Δ: First Approval. BioDrugs 2022, 36, 667–672. [Google Scholar] [CrossRef]
- Mondal, M.; Guo, J.; He, P.; Zhou, D. Recent advances of oncolytic virus in cancer therapy. Hum. Vaccin. Immunother. 2020, 16, 2389–2402. [Google Scholar] [CrossRef]
- Goradel, N.H.; Alizadeh, A.; Hosseinzadeh, S.; Taghipour, M.; Ghesmati, Z.; Arashkia, A.; Negahdari, B. Oncolytic virotherapy as promising immunotherapy against cancer: Mechanisms of resistance to oncolytic viruses. Futur. Oncol. 2022, 18, 245–259. [Google Scholar] [CrossRef]
- Feola, S.; Russo, S.; Ylösmäki, E.; Cerullo, V. Oncolytic ImmunoViroTherapy: A long history of crosstalk between viruses and immune system for cancer treatment. Pharmacol. Ther. 2022, 236, 108103. [Google Scholar] [CrossRef]
- Truong, C.-S.; Yoo, S.Y. Oncolytic Vaccinia Virus in Lung Cancer Vaccines. Vaccines 2022, 10, 240. [Google Scholar] [CrossRef]
- Samulski, R.J.; Zhu, X.; Xiao, X.; Brook, J.D.; Housman, D.E.; Epstein, N.; Hunter, L.A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991, 10, 3941–3950. [Google Scholar] [CrossRef]
- Spunde, K.; Korotkaja, K.; Zajakina, A. Recombinant Viral Vectors for Therapeutic Programming of Tumour Microenvironment: Advantages and Limitations. Biomedicines 2022, 10, 2142. [Google Scholar] [CrossRef]
- Sasso, E.; D’Alise, A.M.; Zambrano, N.; Scarselli, E.; Folgori, A.; Nicosia, A. New viral vectors for infectious diseases and cancer. Semin. Immunol. 2020, 50, 101430. [Google Scholar] [CrossRef]
- Palatini, U.; Miesen, P.; Carballar-Lejarazu, R.; Ometto, L.; Rizzo, E.; Tu, Z.; van Rij, R.P.; Bonizzoni, M. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genom. 2017, 18, 512. [Google Scholar] [CrossRef]
- Buijs, P.R.A.; Verhagen, J.H.E.; van Eijck, C.H.J.; van den Hoogen, B.G. Oncolytic viruses: From bench to bedside with a focus on safety. Hum. Vaccin. Immunother. 2015, 11, 1573–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricca, J.M.; Oseledchyk, A.; Walther, T.; Liu, C.; Mangarin, L.; Merghoub, T.; Wolchok, J.D.; Zamarin, D. Pre-existing Immunity to Oncolytic Virus Potentiates Its Immunotherapeutic Efficacy. Mol. Ther. 2018, 26, 1008–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conti, M. Boosting effect of pre-existing immunity on anti-cancer immunotherapies. Front. Drug Chem. Clin. Res. 2021, 4, 1–6. [Google Scholar] [CrossRef]
- Hromic-Jahjefendic, A.; Lundstrom, K. Viral Vector-Based Melanoma Gene Therapy. Biomedicines 2020, 8, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderplasschen, A.; Mathew, E.; Hollinshead, M.; Sim, R.B.; Smith, G.L. Extracellular enveloped vaccinia virus is resistant to complement because of incorporation of host complement control proteins into its envelope. Proc. Natl. Acad. Sci. USA 1998, 95, 7544–7549. [Google Scholar] [CrossRef] [Green Version]
- Balachandran, S.; Barber, G.N. Vesicular Stomatitis Virus (VSV) Therapy of Tumors. IUBMB Life 2000, 50, 135–138. [Google Scholar] [CrossRef]
- Jayawardena, N.; Poirier, J.T.; Burga, L.N.; Bostina, M. Virus–Receptor Interactions and Virus Neutralization: Insights for Oncolytic Virus Development. Oncolytic Virotherapy 2020, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Muharemagic, D.; Labib, M.; Ghobadloo, S.M.; Zamay, A.S.; Bell, J.C.; Berezovski, M.V. Anti-Fab aptamers for shielding virus from neutralizing antibodies. J. Am. Chem. Soc. 2012, 134, 17168–17177. [Google Scholar] [CrossRef]
- Muharemagic, D.; Zamay, A.; Ghobadloo, S.M.; Evgin, L.; Savitskaya, A.; Bell, J.C.; Berezovski, M.V. Aptamer-facilitated Protection of Oncolytic Virus from Neutralizing Antibodies. Mol. Ther. Nucleic Acids 2014, 3, e167. [Google Scholar] [CrossRef]
- Kumar Kulabhusan, P.; Hussain, B.; Yüce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide Aptamers: New Tools for Targeted Cancer Therapy. Mol. Ther. Nucleic Acids 2014, 3, e182. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, N.; Chan, C.-W.; Lu, A.; Yu, Y.; Zhang, G.; Ren, K. The Application of Microfluidic Technologies in Aptamer Selection. Front. Cell Dev. Biol. 2021, 9, 2548. [Google Scholar] [CrossRef]
- Mironov, V.; Shchugoreva, I.A.; Artyushenko, P.V.; Morozov, D.; Borbone, N.; Oliviero, G.; Zamay, T.N.; Moryachkov, R.V.; Kolovskaya, O.S.; Lukyanenko, K.A.; et al. Structure- and Interaction-Based Design of Anti-SARS-CoV-2 Aptamers. Chem. A Eur. J. 2022, 28, e202104481. [Google Scholar] [CrossRef]
- Bruno, J.G. Applications in Which Aptamers Are Needed or Wanted in Diagnostics and Therapeutics. Pharmaceuticals 2022, 15, 693. [Google Scholar] [CrossRef]
- Yüce, M.; Ullah, N.; Budak, H. Trends in aptamer selection methods and applications. Analyst 2015, 140, 5379–5399. [Google Scholar] [CrossRef]
- Le, A.T.H.; Krylova, S.M.; Kanoatov, M.; Desai, S.; Krylov, S.N. Ideal-Filter Capillary Electrophoresis (IFCE) Facilitates the One-Step Selection of Aptamers. Angew. Chemie Int. Ed. 2019, 58, 2739–2743. [Google Scholar] [CrossRef]
- Dembowski, S.K.; Bowser, M.T. Microfluidic methods for aptamer selection and characterization. Analyst 2018, 143, 21–32. [Google Scholar] [CrossRef]
- Macdonald, J.; Denoyer, D.; Henri, J.; Jamieson, A.; Burvenich, I.J.G.; Pouliot, N.; Shigdar, S. Bifunctional Aptamer–Doxorubicin Conjugate Crosses the Blood–Brain Barrier and Selectively Delivers Its Payload to EpCAM-Positive Tumor Cells. Nucleic Acid Ther. 2020, 30, 117–128. [Google Scholar] [CrossRef]
- Bohrmann, L.; Burghardt, T.; Haynes, C.; Saatchi, K.; Häfeli, U.O. Aptamers used for molecular imaging and theranostics-recent developments. Theranostics 2022, 12, 4010–4050. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Affinito, A.; Quintavalle, C.; Esposito, C.L.; Roscigno, G.; Vilardo, C.; Nuzzo, S.; Ricci-Vitiani, L.; De Luca, G.; Pallini, R.; Kichkailo, A.S.; et al. The Discovery of RNA Aptamers that Selectively Bind Glioblastoma Stem Cells. Mol. Ther.-Nucleic Acids 2019, 18, 99–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, X.; Castanares, M.; Mukherjee, A.; Lupold, S.E. Nucleic Acid Aptamers: Clinical Applications and Promising New Horizons. Curr. Med. Chem. 2011, 18, 4206–4214. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Hussain, A.; Fahimi, H.; Aliakbari, F.; Haj Bloukh, S.; Edis, Z.; Mahdi Nejadi Babadaei, M.; Izadi, Z.; Shiri Varnamkhasti, B.; Jahanshahi, F.; et al. A review on the therapeutic applications of aptamers and aptamer-conjugated nanoparticles in cancer, inflammatory and viral diseases. Arab. J. Chem. 2022, 15, 103626. [Google Scholar] [CrossRef]
- Liu, M.; Wang, L.; Lo, Y.; Shiu, S.C.-C.; Kinghorn, A.B.; Tanner, J.A. Aptamer-Enabled Nanomaterials for Therapeutics, Drug Targeting and Imaging. Cells 2022, 11, 159. [Google Scholar] [CrossRef]
- Kumar, V.; Raj, S.B.; Kanakaraj, L.; Paul, A.D.; Kavitha, K.; Ravi, M.; Sucharitha, P. Aptamer: A Review on It’s In Vitro Selection and Drug Delivery System. Int. J. Appl. Pharm. 2022, 14, 35–42. [Google Scholar] [CrossRef]
- Sousa, D.A.; Carneiro, M.; Ferreira, D.; Moreira, F.T.C.; Sales, M.G.F.; Rodrigues, L.R. Recent Advances in the Selection of Cancer-Specific Aptamers for the Development of Biosensors. Curr. Med. Chem. 2022, 29, 5850–5880. [Google Scholar] [CrossRef]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 2020, 155, 1420–1431. [Google Scholar] [CrossRef]
- Hoellenriegel, J.; Zboralski, D.; Maasch, C.; Rosin, N.Y.; Wierda, W.G.; Keating, M.J.; Kruschinski, A.; Burger, J.A. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood 2014, 123, 1032–1039. [Google Scholar] [CrossRef]
- Zamay, T.N.; Kolovskaya, O.S.; Glazyrin, Y.E.; Zamay, G.S.; Kuznetsova, S.A.; Spivak, E.A.; Wehbe, M.; Savitskaya, A.G.; Zubkova, O.A.; Kadkina, A.; et al. DNA-Aptamer Targeting Vimentin for Tumor Therapy In Vivo. Nucleic Acid Ther. 2014, 24, 160–170. [Google Scholar] [CrossRef]
- Subramanian, N.; Raghunathan, V.; Kanwar, J.R.; Kanwar, R.K.; Elchuri, S.V.; Khetan, V.; Krishnakumar, S. Target-specific delivery of doxorubicin to retinoblastoma using epithelial cell adhesion molecule aptamer. Mol. Vis. 2012, 18, 2783. [Google Scholar]
- Zamay, T.N.; Starkov, A.K.; Kolovskaya, O.S.; Zamay, G.S.; Veprintsev, D.V.; Luzan, N.; Nikolaeva, E.D.; Lukyanenko, K.A.; Artyushenko, P.V.; Shchugoreva, I.A.; et al. Nucleic Acid Aptamers Increase the Anticancer Efficiency and Reduce the Toxicity of Cisplatin-Arabinogalactan Conjugates In Vivo. Nucleic Acid Ther. 2022, 32, 497–506. [Google Scholar] [CrossRef]
- Dassie, J.P.; Liu, X.; Thomas, G.S.; Whitaker, R.M.; Thiel, K.W.; Stockdale, K.R.; Meyerholz, D.K.; McCaffrey, A.P.; McNamara, J.O.; Giangrande, P.H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009, 27, 839–846. [Google Scholar] [CrossRef] [Green Version]
- Esposito, C.; Catuogno, S.; Condorelli, G.; Ungaro, P.; de Franciscis, V. Aptamer Chimeras for Therapeutic Delivery: The Challenging Perspectives. Genes 2018, 9, 529. [Google Scholar] [CrossRef] [Green Version]
- Orava, E.W.; Cicmil, N.; Gariépy, J. Delivering cargoes into cancer cells using DNA aptamers targeting internalized surface portals. Biochim. Biophys. Acta-Biomembr. 2010, 1798, 2190–2200. [Google Scholar] [CrossRef] [Green Version]
- Liechty, W.B.; Kryscio, D.R.; Slaughter, B.V.; Peppas, N.A. Polymers for Drug Delivery Systems. Annu. Rev. Chem. Biomol. Eng. 2010, 1, 149–173. [Google Scholar] [CrossRef] [Green Version]
- Farokhzad, O.C.; Jon, S.; Khademhosseini, A.; Tran, T.-N.T.; LaVan, D.A.; Langer, R. Nanoparticle-Aptamer Bioconjugates. Cancer Res. 2004, 64, 7668–7672. [Google Scholar] [CrossRef] [Green Version]
- Alibolandi, M.; Ramezani, M.; Sadeghi, F.; Abnous, K.; Hadizadeh, F. Epithelial cell adhesion molecule aptamer conjugated PEG–PLGA nanopolymersomes for targeted delivery of doxorubicin to human breast adenocarcinoma cell line in vitro. Int. J. Pharm. 2015, 479, 241–251. [Google Scholar] [CrossRef]
- Xie, X.; Li, F.; Zhang, H.; Lu, Y.; Lian, S.; Lin, H.; Gao, Y.; Jia, L. EpCAM aptamer-functionalized mesoporous silica nanoparticles for efficient colon cancer cell-targeted drug delivery. Eur. J. Pharm. Sci. 2016, 83, 28–35. [Google Scholar] [CrossRef]
- Mongelard, F.; Bouvet, P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr. Opin. Mol. Ther. 2010, 12, 107–114. [Google Scholar]
- Guo, J.; Gao, X.; Su, L.; Xia, H.; Gu, G.; Pang, Z.; Jiang, X.; Yao, L.; Chen, J.; Chen, H. Aptamer-functionalized PEG–PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials 2011, 32, 8010–8020. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Aravind, A.; Nair, R.; Raveendran, S.; Veeranarayanan, S.; Nagaoka, Y.; Fukuda, T.; Hasumura, T.; Morimoto, H.; Yoshida, Y.; Maekawa, T.; et al. Aptamer conjugated paclitaxel and magnetic fluid loaded fluorescently tagged PLGA nanoparticles for targeted cancer therapy. J. Magn. Magn. Mater. 2013, 344, 116–123. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, R.; Fang, X.; Chen, F.; Wang, Y.; Chen, M. Nucleolin targeting AS1411 aptamer modified pH-sensitive micelles for enhanced delivery and antitumor efficacy of paclitaxel. Nano Res. 2015, 8, 201–218. [Google Scholar] [CrossRef]
- Bagalkot, V.; Farokhzad, O.C.; Langer, R.; Jon, S. An Aptamer–Doxorubicin Physical Conjugate as a Novel Targeted Drug-Delivery Platform. Angew. Chemie Int. Ed. 2006, 45, 8149–8152. [Google Scholar] [CrossRef]
- Hu, Y.; Duan, J.; Zhan, Q.; Wang, F.; Lu, X.; Yang, X.-D. Novel MUC1 Aptamer Selectively Delivers Cytotoxic Agent to Cancer Cells In Vitro. PLoS ONE 2012, 7, e31970. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Ga, L.; Aodeng, G.; Wang, Y.; Ai, J. Aptamer-drug conjugates: New probes for imaging and targeted therapy. Biosens. Bioelectron. X 2022, 10, 100126. [Google Scholar] [CrossRef]
- Geng, Z.; Cao, Z.; Liu, R.; Liu, K.; Liu, J.; Tan, W. Aptamer-assisted tumor localization of bacteria for enhanced biotherapy. Nat. Commun. 2021, 12, 6584. [Google Scholar] [CrossRef]
- Yüce, M.; Kurt, H.; Hussain, B.; Ow-Yang, C.W.; Budak, H. Exploiting Stokes and anti-Stokes type emission profiles of aptamer-functionalized luminescent nanoprobes for multiplex sensing applications. ChemistrySelect 2018, 3, 5814–5823. [Google Scholar] [CrossRef]
- Borghei, Y.-S.; Hosseini, M.; Dadmehr, M.; Hosseinkhani, S.; Ganjali, M.R.; Sheikhnejad, R. Visual detection of cancer cells by colorimetric aptasensor based on aggregation of gold nanoparticles induced by DNA hybridization. Anal. Chim. Acta 2016, 904, 92–97. [Google Scholar] [CrossRef]
- Mercier, M.-C.; Dontenwill, M.; Choulier, L. Selection of Nucleic Acid Aptamers Targeting Tumor Cell-Surface Protein Biomarkers. Cancers 2017, 9, 69. [Google Scholar] [CrossRef] [Green Version]
- Schmohl, J.; Vallera, D. CD133, Selectively Targeting the Root of Cancer. Toxins 2016, 8, 165. [Google Scholar] [CrossRef]
- Shabalina, A.V.; Sharko, D.O.; Glazyrin, Y.E.; Bolshevich, E.A.; Dubinina, O.V.; Kim, A.M.; Veprintsev, D.V.; Lapin, I.N.; Zamay, G.S.; Krat, A.V.; et al. Development of Electrochemical Aptasensor for Lung Cancer Diagnostics in Human Blood. Sensors 2021, 21, 7851. [Google Scholar] [CrossRef]
- Ruiz Ciancio, D.; Vargas, M.; Thiel, W.; Bruno, M.; Giangrande, P.; Mestre, M. Aptamers as Diagnostic Tools in Cancer. Pharmaceuticals 2018, 11, 86. [Google Scholar] [CrossRef] [Green Version]
- Molefe, P.; Masamba, P.; Oyinloye, B.; Mbatha, L.; Meyer, M.; Kappo, A. Molecular Application of Aptamers in the Diagnosis and Treatment of Cancer and Communicable Diseases. Pharmaceuticals 2018, 11, 93. [Google Scholar] [CrossRef]
- Bognár, Z.; Gyurcsányi, R.E. Aptamers against Immunoglobulins: Design, Selection and Bioanalytical Applications. Int. J. Mol. Sci. 2020, 21, 5748. [Google Scholar] [CrossRef]
- Ghobadloo, S.M.; Gargaun, A.; Casselman, R.; Muharemagic, D.; Berezovski, M.V. Aptamer-Facilitated Cryoprotection of Viruses. ACS Med. Chem. Lett. 2014, 5, 1240–1244. [Google Scholar] [CrossRef] [Green Version]
- Tong, G.J.; Hsiao, S.C.; Carrico, Z.M.; Francis, M.B. Viral Capsid DNA Aptamer Conjugates as Multivalent Cell-Targeting Vehicles. J. Am. Chem. Soc. 2009, 131, 11174–11178. [Google Scholar] [CrossRef] [Green Version]
- Lau, J.L.; Baksh, M.M.; Fiedler, J.D.; Brown, S.D.; Kussrow, A.; Bornhop, D.J.; Ordoukhanian, P.; Finn, M.G. Evolution and Protein Packaging of Small-Molecule RNA Aptamers. ACS Nano 2011, 5, 7722–7729. [Google Scholar] [CrossRef]
- Wu, Z.; Tang, L.J.; Zhang, X.B.; Jiang, J.H.; Tan, W. Aptamer-modified nanodrug delivery systems. ACS Nano 2011, 5, 7696–7699. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Liu, S.; Song, T.; Chen, F.; Zhang, J.; Huang, J.; Wan, S.; Lu, Y.; Chen, H.; Tan, W.; et al. Spherical Neutralizing Aptamer Inhibits SARS-CoV-2 Infection and Suppresses Mutational Escape. J. Am. Chem. Soc. 2021, 143, 21541–21548. [Google Scholar] [CrossRef]
- Sun, M.; Liu, S.; Wei, X.; Wan, S.; Huang, M.; Song, T.; Lu, Y.; Weng, X.; Lin, Z.; Chen, H.; et al. Aptamer Blocking Strategy Inhibits SARS-CoV-2 Virus Infection. Angew. Chemie Int. Ed. 2021, 60, 10266–10272. [Google Scholar] [CrossRef] [PubMed]
- Abrego-Martinez, J.C.; Jafari, M.; Chergui, S.; Pavel, C.; Che, D.; Siaj, M. Aptamer-based electrochemical biosensor for rapid detection of SARS-CoV-2: Nanoscale electrode-aptamer-SARS-CoV-2 imaging by photo-induced force microscopy. Biosens. Bioelectron. 2022, 195, 113595. [Google Scholar] [CrossRef] [PubMed]
- Hubálek, Z. Protectants used in the cryopreservation of microorganisms. Cryobiology 2003, 46, 205–229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pritchard, E.; Hu, X.; Valentin, T.; Panilaitis, B.; Omenetto, F.G.; Kaplan, D.L. Stabilization of vaccines and antibiotics in silk and eliminating the cold chain. Proc. Natl. Acad. Sci. USA 2012, 109, 11981–11986. [Google Scholar] [CrossRef] [Green Version]
- Marton, H.L.; Styles, K.M.; Kilbride, P.; Sagona, A.P.; Gibson, M.I. Polymer-Mediated Cryopreservation of Bacteriophages. Biomacromolecules 2021, 22, 5281–5289. [Google Scholar] [CrossRef]
- Nakanishi, K.; Tomita, M.; Tsumoto, K. Membrane fusion and infection abilities of baculovirus virions are preserved during freezing and thawing in the presence of trehalose. Biosci. Biotechnol. Biochem. 2020, 84, 686–694. [Google Scholar] [CrossRef]
- Pradhan, S.; Varsani, A.; Leff, C.; Swanson, C.J.; Hariadi, R.F. Viral Aggregation: The Knowns and Unknowns. Viruses 2022, 14, 438. [Google Scholar] [CrossRef]
- Gerba, C.P.; Betancourt, W.Q. Viral Aggregation: Impact on Virus Behavior in the Environment. Environ. Sci. Technol. 2017, 51, 7318–7325. [Google Scholar] [CrossRef]



| № | The Name of Aptamers | Virus | Targets | Mechanisms | References |
|---|---|---|---|---|---|
| 1 | 41-nucleotide DNA aptamer | Bacteriophage MS2 | Tyrosine kinase receptor on Jurkat T cells | cell binding, endocytosis, and translocation to lysosomes for degradation | [99] |
| 2 | RNA aptamer | VLPs from bacteriophage Qβ | heteroaryldihydropyrimidine structure | development of encapsulation technique | [100] |
| 3 | DNA aptamers | VSV | Fab of anti–VSV pAbs | shielding virus from nAbs | [49] |
| 4 | Tetramer of DNA aptamers | VSV | Fab of anti–VSV pAbs, VSVs | shielding the virus from nAbs, increasing infectivity | [50] |
| 5 | Quadramer of DNA aptamers | VSV | Fab of anti–VSV pAbs | Prevention of virus aggregation, protection against nAbs, cryoprotection | [98] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dymova, M.A.; Kichkailo, A.S.; Kuligina, E.V.; Richter, V.A. Aptamers Enhance Oncolytic Viruses’ Antitumor Efficacy. Pharmaceutics 2023, 15, 151. https://doi.org/10.3390/pharmaceutics15010151
Dymova MA, Kichkailo AS, Kuligina EV, Richter VA. Aptamers Enhance Oncolytic Viruses’ Antitumor Efficacy. Pharmaceutics. 2023; 15(1):151. https://doi.org/10.3390/pharmaceutics15010151
Chicago/Turabian StyleDymova, Maya A., Anna S. Kichkailo, Elena V. Kuligina, and Vladimir A. Richter. 2023. "Aptamers Enhance Oncolytic Viruses’ Antitumor Efficacy" Pharmaceutics 15, no. 1: 151. https://doi.org/10.3390/pharmaceutics15010151