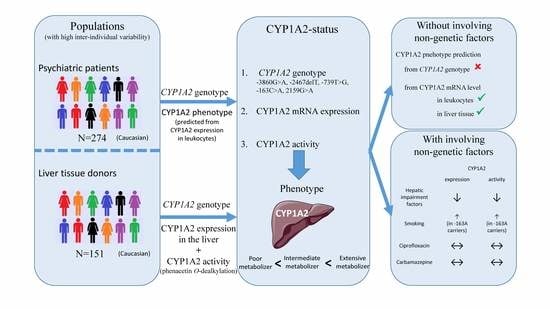
CYP1A2 mRNA Expression Rather than Genetic Variants Indicate Hepatic CYP1A2 Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Liver Microsomes and RNA Samples
2.2. CYP1A2 Enzyme Assay
2.3. CYP1A2 Genotyping
2.4. Analysis of CYP1A2 mRNA Levels by Quantitative Real-Time PCR
2.5. Statistical Analysis
3. Results
3.1. CYP1A2 Haplotypes and Genotypes of Liver Tissue Donors and Psychiatric Patients
3.2. Hepatic CYP1A2 Activities and mRNA Expression
3.3. Impact of −163C>A (rs762551) on CYP1A2 Activity and mRNA Expression in the Liver
3.4. Impact of Genetic and Non-Genetic Factors on Inter-Individual Variation in Hepatic CYP1A2 Activities and Expression
3.5. Impact of Genetic and Non-Genetic Factors on CYP1A2 mRNA Expression in Psychiatric Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shimada, T.; Yamazaki, H.; Mimura, M.; Inui, Y.; Guengerich, F.P. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: Studies with liver microsomes of 30 Japanese and 30 Caucasians. J. Pharmacol. Exp. Ther. 1994, 270, 414–423. [Google Scholar] [PubMed]
- Mercurio, M.G.; Shiff, S.J.; Galbraith, R.A.; Sassa, S. Expression of cytochrome P450 mRNAs in the colon and the rectum in normal human subjects. Biochem. Biophys. Res. Commun. 1995, 210, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, M.; Nishimura, M.; Ogino, D.; Chiba, R.; Ikai, I.; Ueda, N.; Naito, S.; Kuribayashi, S.; Moustafa, M.A.; Uchida, T.; et al. Cytochrome P450 gene expression levels in peripheral blood mononuclear cells in comparison with the liver. Cancer Sci. 2004, 95, 520–529. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.; Caccavale, R.J.; Kehoe, J.J.; Thomas, P.E.; Iba, M.M. CYP1A2 is expressed along with CYP1A1 in the human lung. Cancer Lett. 2001, 171, 113–120. [Google Scholar] [CrossRef]
- Wang, B.; Zhou, S.-F. Synthetic and natural compounds that interact with human cytochrome P450 1A2 and implications in drug development. Curr. Med. Chem. 2009, 16, 4066–4218. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.-F.; Yang, L.-P.; Wei, M.; Duan, W.; Chan, E. Insights into the structure, function, and regulation of human cytochrome P450 1A2. Curr. Drug Metab. 2009, 10, 713–729. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.-F.; Wang, B.; Yang, L.-P.; Liu, J.-P. Structure, function, regulation and polymorphism and the clinical significance of human cytochrome P450 1A2. Drug Metab. Rev. 2010, 42, 268–354. [Google Scholar] [CrossRef]
- Jorge-Nebert, L.F.; Jiang, Z.; Chakraborty, R.; Watson, J.; Jin, L.; McGarvey, S.T.; Deka, R.; Nebert, D.W. Analysis of human CYP1A1 and CYP1A2 genes and their shared bidirectional promoter in eight world populations. Hum. Mutat. 2010, 31, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Ueda, R.; Iketaki, H.; Nagata, K.; Kimura, S.; Gonzalez, F.J.; Kusano, K.; Yoshimura, T.; Yamazoe, Y. A common regulatory region functions bidirectionally in transcriptional activation of the human CYP1A1 and CYP1A2 genes. Mol. Pharmacol. 2006, 69, 1924–1930. [Google Scholar] [CrossRef] [Green Version]
- Klein, K.; Winter, S.; Turpeinen, M.; Schwab, M.; Zanger, U.M. Pathway-targeted pharmacogenomics of CYP1A2 in human liver. Front. Pharmacol. 2010, 1, 129. [Google Scholar] [CrossRef] [Green Version]
- Monostory, K.; Pascussi, J.-M.; Kóbori, L.; Dvorak, Z. Hormonal regulation of CYP1A expression. Drug Metab. Rev. 2009, 41, 547–572. [Google Scholar] [CrossRef]
- Saruwatari, J.; Nakagawa, K.; Shindo, J.; Tajiri, T.; Fujieda, M.; Yamazaki, H.; Kamataki, T.; Ishizaki, T. A population phenotyping study of three drug-metabolizing enzymes in Kyushu, Japan, with use of the caffeine test. Clin. Pharmacol. Ther. 2002, 72, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P.; Parikh, A.; Turesky, R.J.; Josephy, P.D. Inter-individual differences in the metabolism of environmental toxicants: Cytochrome P450 1A2 as a prototype. Mutat. Res.—Fundam. Mol. Mech. Mutagen. 1999, 428, 115–124. [Google Scholar] [CrossRef]
- Gunes, A.; Dahl, M.-L. Variation in CYP1A2 activity and its clinical implications: Influence of environmental factors and genetic polymorphisms. Pharmacogenomics 2008, 9, 625–637. [Google Scholar] [CrossRef]
- Han, X.-M.; Ouyang, D.-S.; Chen, X.-P.; Shu, Y.; Jiang, C.-H.; Tan, Z.-R.; Zhou, H.-H. Inducibility of CYP1A2 by omeprazole in vivo related to the genetic polymorphism of CYP1A2. Br. J. Clin. Pharmacol. 2002, 54, 540–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobrinas, M.; Cornuz, J.; Oneda, B.; Kohler Serra, M.; Puhl, M.; Eap, C.B. Impact of smoking, smoking cessation, and genetic polymorphisms on CYP1A2 activity and inducibility. Clin. Pharmacol. Ther. 2011, 90, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Sachse, C.; Brockmöller, J.; Bauer, S.; Roots, I. Functional significance of a C→A polymorphism in intron 1 of the cytochrome P450 CYP1A2 gene tested with caffeine. Br. J. Clin. Pharmacol. 1999, 47, 445–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kootstra-Ros, J.E.; Smallegoor, W.; van der Weide, J. The cytochrome P450 CYP1A2 genetic polymorphisms *1F and *1D do not affect clozapine clearance in a group of schizophrenic patients. Ann. Clin. Biochem. Int. J. Lab. Med. 2005, 42, 216–219. [Google Scholar] [CrossRef] [Green Version]
- Laika, B.; Leucht, S.; Heres, S.; Schneider, H.; Steimer, W. Pharmacogenetics and olanzapine treatment: CYP1A2*1F and serotonergic polymorphisms influence therapeutic outcome. Pharm. J. 2010, 10, 20–29. [Google Scholar] [CrossRef]
- Ghotbi, R.; Christensen, M.; Roh, H.-K.; Ingelman-Sundberg, M.; Aklillu, E.; Bertilsson, L. Comparisons of CYP1A2 genetic polymorphisms, enzyme activity and the genotype-phenotype relationship in Swedes and Koreans. Eur. J. Clin. Pharmacol. 2007, 63, 537–546. [Google Scholar] [CrossRef]
- Na Takuathung, M.; Hanprasertpong, N.; Teekachunhatean, S.; Koonrungsesomboon, N. Impact of CYP1A2 genetic polymorphisms on pharmacokinetics of antipsychotic drugs: A systematic review and meta-analysis. Acta Psychiatr. Scand. 2019, 139, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Thorn, C.F.; Aklillu, E.; Klein, T.E.; Altman, R.B. PharmGKB summary: Very important pharmacogene information for CYP1A2. Pharmacogenet. Genomics 2012, 22, 73–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soyama, A.; Saito, Y.; Hanioka, N.; Maekawa, K.; Komamura, K.; Kamakura, S.; Kitakaze, M.; Tomoike, H.; Ueno, K.; Goto, Y.; et al. Single nucleotide polymorphisms and haplotypes of CYP1A2 in a Japanese population. Drug Metab. Pharmacokinet. 2005, 20, 24–33. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, M.; Yokoi, T.; Mizutani, M.; Kinoshitah, M.; Funayama, M.; Kamataki, T. Genetic polymorphism in the 5’-flanking region of human CYP1A2 gene: Effect on the CYP1A2 inducibility in humans. J. Biochem. 1999, 125, 803–808. [Google Scholar] [CrossRef]
- Djordjevic, N.; Ghotbi, R.; Jankovic, S.; Aklillu, E. Induction of CYP1A2 by heavy coffee consumption is associated with the CYP1A2 −163C>A polymorphism. Eur. J. Clin. Pharmacol. 2010, 66, 697–703. [Google Scholar] [CrossRef]
- Aklillu, E.; Carrillo, J.A.; Makonnen, E.; Hellman, K.; Pitarque, M.; Bertilsson, L.; Ingelman-Sundberg, M. Genetic polymorphism of CYP1A2 in Ethiopians affecting induction and expression: Characterization of novel haplotypes with single-nucleotide polymorphisms in intron 1. Mol. Pharmacol. 2003, 64, 659–669. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, E. Gender-related differences in pharmacokinetics and their clinical significance. J. Clin. Pharm. Ther. 1999, 24, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, A.; Mudra, D.R.; Johnson, C.; Dwyer, A.; Carroll, K.M. The effects of gender, age, ethnicity, and liver cirrhosis on cytochrome P450 enzyme activity in human liver microsomes and inducibility in cultured human hepatocytes. Toxicol. Appl. Pharmacol. 2004, 199, 193–209. [Google Scholar] [CrossRef]
- Ou-Yang, D.-S.; Huang, S.-L.; Wang, W.; Xie, H.-G.; Xu, Z.-H.; Shu, Y.; Zhou, H.-H. Phenotypic polymorphism and gender-related differences of CYP1A2 activity in a Chinese population. Br. J. Clin. Pharmacol. 2000, 49, 145–151. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y.-F.; Corton, J.C.; Klaassen, C.D. Expression of cytochrome P450 isozyme transcripts and activities in human livers. Xenobiotica 2021, 51, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Denison, M.S.; Nagy, S.R. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003, 43, 309–334. [Google Scholar] [CrossRef] [PubMed]
- Perera, V.; Gross, A.S.; McLachlan, A.J. Influence of environmental and genetic factors on CYP1A2 activity in individuals of South Asian and European ancestry. Clin. Pharmacol. Ther. 2012, 92, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Hiemke, C.; Pfuhlmann, B. Interactions and Monitoring of Antipsychotic Drugs. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 212, pp. 241–265. ISBN 9783642257605. [Google Scholar]
- Brewer, L.; Williams, D. Clinically relevant drug-drug and drug-food interactions. Pharmaceut. Med. 2013, 27, 9–23. [Google Scholar] [CrossRef]
- Granfors, M.T.; Backman, J.T.; Laitila, J.; Neuvonen, P.J. Oral contraceptives containing ethinyl estradiol and gestodene markedly increase plasma concentrations and effects of tizanidine by inhibiting cytochrome P450 1A2. Clin. Pharmacol. Ther. 2005, 78, 400–411. [Google Scholar] [CrossRef]
- Van der Hoeven, T.A.; Coon, M.J. Preparation and properties of partially purified cytochrome P-450 and reduced nicotinamide adenine dinucleotide phosphate-cytochrome P-450 reductase from rabbit liver microsomes. J. Biol. Chem. 1974, 249, 6302–6310. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Butler, M.A.; Iwasaki, M.; Guengerich, F.P.; Kadlubar, F.F. Human cytochrome P-450PA (P-450IA2), the phenacetin O-deethylase, is primarily responsible for the hepatic 3-demethylation of caffeine and N-oxidation of carcinogenic arylamines. Proc. Natl. Acad. Sci. USA 1989, 86, 7696–7700. [Google Scholar] [CrossRef] [Green Version]
- Déri, M.T.; Kiss, Á.F.; Tóth, K.; Paulik, J.; Sárváry, E.; Kóbori, L.; Monostory, K. End-stage renal disease reduces the expression of drug-metabolizing cytochrome P450s. Pharmacol. Rep. 2020, 72, 1695–1705. [Google Scholar] [CrossRef]
- Stephens, M.; Smith, N.J.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef] [Green Version]
- Stephens, M.; Scheet, P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005, 76, 449–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevalier, D.; Cauffiez, C.; Allorge, D.; Lo-Guidice, J.M.; Lhermitte, M.; Lafitte, J.J.; Broly, F. Five novel natural allelic variants—951A>C, 1042G>A (D348N), 1156A>T (I386F), 1217G>A (C406Y) and 1291C>T (C431Y)—of the human CYP1A2 gene in a French Caucasian population. Hum. Mutat. 2001, 17, 355. [Google Scholar] [CrossRef] [PubMed]
- Perera, V.; Gross, A.S.; Polasek, T.M.; Qin, Y.; Rao, G.; Forrest, A.; Xu, J.; McLachlan, A.J. Considering CYP1A2 phenotype and genotype for optimizing the dose of olanzapine in the management of schizophrenia. Expert Opin. Drug Metab. Toxicol. 2013, 9, 1115–1137. [Google Scholar] [CrossRef] [PubMed]
- Temesvári, M.; Kóbori, L.; Paulik, J.; Saŕvaŕy, E.; Belic, A.; Monostory, K. Estimation of drug-metabolizing capacity by cytochrome P450 genotyping and expression. J. Pharmacol. Exp. Ther. 2012, 341, 294–305. [Google Scholar] [CrossRef] [Green Version]
- Chung, W.G.; Kang, J.H.; Park, C.S.; Cho, M.H.; Cha, Y.N. Effect of age and smoking on in vivo CYP1A2, flavin-containing monooxygenase, and xanthine oxidase activities in Koreans: Determination by caffeine metabolism. Clin. Pharmacol. Ther. 2000, 67, 258–266. [Google Scholar] [CrossRef]
- Granfors, M.T.; Backman, J.T.; Neuvonen, M.; Neuvonen, P.J. Ciprofloxacin greatly increases concentrations and hypotensive effect of tizanidine by inhibiting its cytochrome P450 1A2-mediated presystemic metabolism. Clin. Pharmacol. Ther. 2004, 76, 598–606. [Google Scholar] [CrossRef]
- Ayano, G. Psychotropic medications metabolized by cytochromes P450 (CYP) 2D6 enzyme and relevant drug interactions. Clin. Pharmacol. Biopharm. 2016, 5, 2–5. [Google Scholar] [CrossRef]
- Nehlig, A. Interindividual differences in caffeine metabolism and factors driving caffeine consumption. Pharmacol. Rev. 2018, 70, 384–411. [Google Scholar] [CrossRef] [Green Version]
- Koonrungsesomboon, N.; Khatsri, R.; Wongchompoo, P.; Teekachunhatean, S. The impact of genetic polymorphisms on CYP1A2 activity in humans: A systematic review and meta-analysis. Pharm. J. 2018, 18, 760–768. [Google Scholar] [CrossRef]
- Ing Lorenzini, K.; Desmeules, J.; Rollason, V.; Bertin, S.; Besson, M.; Daali, Y.; Samer, C.F. CYP450 genotype—Phenotype concordance using the geneva micrococktail in a clinical setting. Front. Pharmacol. 2021, 12, 730637. [Google Scholar] [CrossRef]
- Rodríguez-Antona, C.; Donato, M.T.; Pareja, E.; Gómez-Lechón, M.-J.; Castell, J.V. Cytochrome P-450 mRNA expression in human liver and its relationship with enzyme activity. Arch. Biochem. Biophys. 2001, 393, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Sachse, C.; Bhambra, U.; Smith, G.; Lightfoot, T.J.; Barrett, J.H.; Scollay, J.; Garner, R.C.; Boobis, A.R.; Wolf, C.R.; Gooderham, N.J. Polymorphisms in the cytochrome P450 CYP1A2 gene (CYP1A2) in colorectal cancer patients and controls: Allele frequencies, linkage disequilibrium and influence on caffeine metabolism. Br. J. Clin. Pharmacol. 2003, 55, 68–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skarke, C.; Kirchhof, A.; Geisslinger, G.; Lötsch, J. Rapid genotyping for relevant CYP1A2 alleles by pyrosequencing. Eur. J. Clin. Pharmacol. 2005, 61, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Pavanello, S.; Pulliero, A.; Lupi, S.; Gregorio, P.; Clonfero, E. Influence of the genetic polymorphism in the 5′-noncoding region of the CYP1A2 gene on CYP1A2 phenotype and urinary mutagenicity in smokers. Mutat. Res.—Genet. Toxicol. Environ. Mutagen. 2005, 587, 59–66. [Google Scholar] [CrossRef]
- Pavanello, S.; Fedeli, U.; Mastrangelo, G.; Rota, F.; Overvad, K.; Raaschou-Nielsen, O.; Tjønneland, A.; Vogel, U. Role of CYP1A2 polymorphisms on lung cancer risk in a prospective study. Cancer Genet. 2012, 205, 278–284. [Google Scholar] [CrossRef]
- Caudle, K.; Klein, T.; Hoffman, J.; Muller, D.; Whirl-Carrillo, M.; Gong, L.; McDonagh, E.; Sangkuhl, K.; Thorn, C.; Schwab, M.; et al. Incorporation of pharmacogenomics into routine clinical practice: The Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr. Drug Metab. 2014, 15, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Ingelman-Sundberg, M.; Lauschke, V.M. Worldwide distribution of cytochrome P450 alleles: A meta-analysis of population-scale sequencing projects. Clin. Pharmacol. Ther. 2017, 102, 688–700. [Google Scholar] [CrossRef] [Green Version]
- Van der Wouden, C.; Cambon-Thomsen, A.; Cecchin, E.; Cheung, K.; Dávila-Fajardo, C.; Deneer, V.; Dolžan, V.; Ingelman-Sundberg, M.; Jönsson, S.; Karlsson, M.; et al. Implementing pharmacogenomics in Europe: Design and implementation strategy of the Ubiquitous Pharmacogenomics Consortium. Clin. Pharmacol. Ther. 2017, 101, 341–358. [Google Scholar] [CrossRef]
- Swen, J.J.; Nijenhuis, M.; van Rhenen, M.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; Mulder, H.; Rongen, G.A.; van Schaik, R.H.N.; van der Weide, J.; et al. Pharmacogenetic information in clinical guidelines: The European perspective. Clin. Pharmacol. Ther. 2018, 103, 795–801. [Google Scholar] [CrossRef]
- Ruan, C.-J.; de Leon, J. Is there a future for CYP1A2 pharmacogenetics in the optimal dosing of clozapine? Pharmacogenomics 2020, 21, 369–373. [Google Scholar] [CrossRef]
- Tian, D.-D.; Natesan, S.; White, J.R.; Paine, M.F. Effects of common CYP1A2 genotypes and other key factors on intraindividual variation in the caffeine metabolic ratio: An exploratory analysis. Clin. Transl. Sci. 2019, 12, 39–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceni, E.; Mello, T.; Galli, A. Pathogenesis of alcoholic liver disease: Role of oxidative metabolism. World J. Gastroenterol. 2014, 20, 17756–17772. [Google Scholar] [CrossRef]
- Lown, K.; Kolars, J.; Turgeon, K.; Merion, R.; Wrighton, S.A.; Watkins, P.B. The erythromycin breath test selectively measures P450IIIA in patients with severe liver disease. Clin. Pharmacol. Ther. 1992, 51, 229–238. [Google Scholar] [CrossRef] [Green Version]
- Seitz, H.K.; Mueller, S. Alcoholic Liver Disease. In Clinical Hepatology; Springer: Berlin/Heidelberg, Germany, 2010; Volume 2, pp. 1111–1151. ISBN 978-3-642-04509-7. [Google Scholar]
- Fontana, R.J.; Shakil, A.O.; Greenson, J.K.; Boyd, I.; Lee, W.M. Acute liver failure due to amoxicillin and amoxicillin/clavulanate. Dig. Dis. Sci. 2005, 50, 1785–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gresser, U. Amoxicillin-clavulanic acid therapy may be associated with severe side effects—Review of the literature. Eur. J. Med. Res. 2001, 6, 139–149. [Google Scholar]
- Fekete, F.; Mangó, K.; Déri, M.; Incze, E.; Minus, A.; Monostory, K. Impact of genetic and non-genetic factors on hepatic CYP2C9 expression and activity in Hungarian subjects. Sci. Rep. 2021, 11, 17081. [Google Scholar] [CrossRef]
- Kiss, Á.F.; Vaskó, D.; Déri, M.T.; Tóth, K.; Monostory, K. Combination of CYP2C19 genotype with non-genetic factors evoking phenoconversion improves phenotype prediction. Pharmacol. Rep. 2018, 70, 525–532. [Google Scholar] [CrossRef] [Green Version]
- Meyer, J.M. Individual changes in clozapine levels after smoking cessation: Results and a predictive model. J. Clin. Psychopharmacol. 2001, 21, 569–574. [Google Scholar] [CrossRef]
- Carrillo, J.A.; Herráiz, A.G.; Ramos, S.I.; Gervasini, G.; Vizcaíno, S.; Benítez, J. Role of the smoking-induced cytochrome P450 (CYP)1A2 and polymorphic CYP2D6 in steady-state concentration of olanzapine. J. Clin. Psychopharmacol. 2003, 23, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Lucas, R.A.; Gilfillan, D.J.; Bergstrom, R.F. A pharmacokinetic interaction between carbamazepine and olanzapine: Observations on possible mechanism. Eur. J. Clin. Pharmacol. 1998, 54, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.-F.; Yang, L.-P.; Zhou, Z.-W.; Liu, Y.-H.; Chan, E. Insights into the substrate specificity, inhibitors, regulation, and polymorphisms and the clinical impact of human cytochrome P450 1A2. AAPS J. 2009, 11, 481–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Liver Tissue Donors | Psychiatric Patients | |||
|---|---|---|---|---|
| Number of subjects | 151 | 274 | ||
| Age (year) 1 | 46 (18; 74) | 39 (18; 76) | ||
| Gender (male/female) | 82/69 | 110/164 | ||
| Cause of death | Psychiatric disorders | |||
| Accident | Car/motor/bike accident | 14 | Schizophrenia | 140 |
| Anoxic cerebral injury/asphyxia | 5 | Persistent delusional disorders | 2 | |
| Seizure induced cerebral injury | 3 | Acute and transient psychotic disorders | 16 | |
| Suicide | 4 | Schizoaffective disorders | 44 | |
| Unknown cerebral injury | 9 | Other non-organic psychotic disorders | 2 | |
| Cerebral hemorrhage/hematoma | Ruptured cerebral aneurysm | 4 | Bipolar affective disorder | 52 |
| Epidural hematoma | 1 | Depressive episode | 5 | |
| Intraventricular hemorrhage | 8 | Recurrent depressive disorder | 3 | |
| Subarachnoid hemorrhage | 31 | Other anxiety disorders | 1 | |
| Subdural hemorrhage | 8 | Specific personality disorders | 2 | |
| Unknown cerebral hemorrhage | 15 | Unknown | 7 | |
| Stroke | Ischemic stroke | 7 | ||
| Hemorrhagic stroke | 2 | |||
| Tumor | 36 | |||
| Unknown | 4 | |||
| Non-genetic factors | ||||
| Amoxicillin+clavulanic acid therapy | 8 | 0 | ||
| Chronic alcohol consumption | 19 | 0 | ||
| Medication with CYP1A2 inducer | 0 | 6 | ||
| Medication with CYP1A2 inhibitor | 1 | 0 | ||
| Smoking | 8 | 96 | ||
| CYP1A2 Allele | Nucleotide Changes | N | Frequency (%) | |||
|---|---|---|---|---|---|---|
| Tissue donors | Psychiatric patients | Tissue donors | Psychiatric patients | Caucasian populations 1 | ||
| *1 2 | None | 99 | 176 | 32.8 | 32.1 | 24.4–63.5 |
| *1C | −3860G>A | 0 | 0 | 0 | 0 | 0.4–4 |
| *1D | −2467delT | 0 | 0 | 0 | 0 | 3.4–11 |
| *1E | −739T>G | 0 | 0 | 0 | 0 | 0.4–6 |
| *1F | −163C>A | 0 | 2 | 0 | 0.4 | 32–57 |
| *1L | −3860G>A; −2467delT; −163C>A; 5347T>C | 6 | 6 | 2.0 | 1.1 | 0.8 |
| *1M | −163C>A; 2159G>A | 181 | 343 | 59.9 | 62.6 | 54.8 |
| *1V | −2467delT; −163C>A | 11 | 15 | 3.6 | 2.7 | 2.8–12.3 |
| *1W | −3113A>G; −2467delT; −739T>G; −163C>A | 5 | 6 | 1.7 | 1.1 | 1.2–2.1 |
| CYP1A2 genotype | N | Frequency (%) | ||||
| Tissue donors | Psychiatric patients | Tissue donors | Psychiatric patients | |||
| *1/*1 | 19 | 35 | 12.6 | 12.8 | ||
| *1/*1F | 0 | 2 | 0 | 0.7 | ||
| *1/*1L | 2 | 0 | 1.3 | 0 | ||
| *1/*1M | 54 | 96 | 35.8 | 35.0 | ||
| *1/*1V | 3 | 5 | 2.0 | 1.8 | ||
| *1/*1W | 2 | 3 | 1.3 | 1.1 | ||
| *1M/*1L | 4 | 4 | 2.6 | 1.5 | ||
| *1M/*1M | 56 | 117 | 37.1 | 42.7 | ||
| *1M/*1V | 8 | 7 | 5.3 | 2.6 | ||
| *1M/*1W | 3 | 2 | 2 | 0.7 | ||
| *1V/*1L | 0 | 1 | 0 | 0.4 | ||
| *1V/*1V | 0 | 1 | 0 | 0.4 | ||
| *1W/*1L | 0 | 1 | 0 | 0.4 | ||
| Variable | CYP1A2 Activity | CYP1A2 mRNA Expression | |||||
|---|---|---|---|---|---|---|---|
| Coefficient B (SE) | Coefficient ß | p Value | Coefficient B (SE) | Coefficient ß | p Value | ||
| SNPs, non-genetic | Constant | 124.92 (54.99) | 0.025 | 0.125 (0.052) | 0.017 | ||
| −3860G>A (rs2069514) | −87.27 (119.26) | −0.077 | 0.466 | 0.014 (0.086) | 0.021 | 0.867 | |
| −163C>A (rs762551) | −2.77 (136.61) | −0.005 | 0.984 | −0.034 (0.098) | −0.070 | 0.728 | |
| −2467delT (rs35694136) | 212.01 (81.90) | 0.312 | 0.011 | 0.164 (0.063) | 0.385 | 0.011 | |
| −739T>G (rs20695) | −288.73 (218.63) | −0.128 | 0.189 | −0.130 (0.100) | −0.154 | 0.198 | |
| 2159G>A (rs2472304) | 70.81 (127.58) | 0.130 | 0.580 | 0.084 (0.088) | 0.194 | 0.341 | |
| Sex | −8.02 (34.82) | −0.020 | 0.818 | −0.023 (0.040) | −0.066 | 0.569 | |
| Age | 78.69 (35.06) | 0.191 | 0.027 | 0.058 (0.038) | 0.166 | 0.128 | |
| Amoxicillin+clavulanic acid therapy | −133.89 (74.94) | −0.154 | 0.077 | −0.161 (0.081) | −0.213 | 0.050 | |
| Chronic alcohol consumption | −170.67 (50.01) | −0.292 | <0.001 | −0.146 (0.049) | −0.338 | 0.004 | |
| Haplotypes, non-genetic | Constant | 124.12 (51.83) | 0.018 | 0.119 (0.046) | 0.011 | ||
| −3860A/−2467delT/−739T/−163A/2159G | 127.91 (95.67) | 0.112 | 0.184 | 0.166 (0.069) | 0.238 | 0.019 | |
| −3860G/−2467T/−739T/−163A/2159A | 68.27 (46.56) | 0.125 | 0.145 | 0.057 (0.044) | 0.132 | 0.199 | |
| −3860G/−2467delT/−739T/−163A/2159G | 211.41 (72.86) | 0.243 | 0.004 | 0.154 (0.058) | 0.267 | 0.009 | |
| −3860G/−2467delT/−739G/−163A/2159G | −79.18 (190.15) | −0.035 | 0.678 | 0.027 (0.082) | 0.032 | 0.743 | |
| Sex | −7.59 (34.35) | −0.019 | 0.825 | −0.021 (0.039) | −0.060 | 0.592 | |
| Age | 79.19 (34.76) | 0.193 | 0.024 | 0.057 (0.037) | 0.163 | 0.127 | |
| Activity reducing factors | −160.33 (43.82) | −0.316 | <0.001 | −0.149 (0.045) | −0.375 | 0.002 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fekete, F.; Mangó, K.; Minus, A.; Tóth, K.; Monostory, K. CYP1A2 mRNA Expression Rather than Genetic Variants Indicate Hepatic CYP1A2 Activity. Pharmaceutics 2022, 14, 532. https://doi.org/10.3390/pharmaceutics14030532
Fekete F, Mangó K, Minus A, Tóth K, Monostory K. CYP1A2 mRNA Expression Rather than Genetic Variants Indicate Hepatic CYP1A2 Activity. Pharmaceutics. 2022; 14(3):532. https://doi.org/10.3390/pharmaceutics14030532
Chicago/Turabian StyleFekete, Ferenc, Katalin Mangó, Annamária Minus, Katalin Tóth, and Katalin Monostory. 2022. "CYP1A2 mRNA Expression Rather than Genetic Variants Indicate Hepatic CYP1A2 Activity" Pharmaceutics 14, no. 3: 532. https://doi.org/10.3390/pharmaceutics14030532