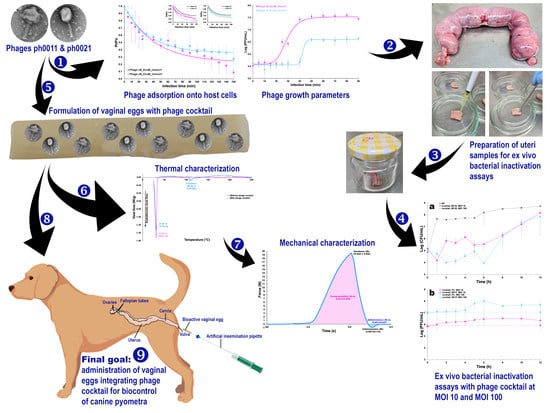
Isolation and Molecular Characterization of Two Novel Lytic Bacteriophages for the Biocontrol of Escherichia coli in Uterine Infections: In Vitro and Ex Vivo Preliminary Studies in Veterinary Medicine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. Chemicals
2.3. Isolation and Characterization of the Bacterial Strains Responsible for Animal Uterine Infections (Pyometra)
2.4. Phage Enrichment, Isolation and Enumeration
2.5. Phage PEG-Precipitation
2.6. UV-Vis Spectral Scans for Determination of Phage Particle Molar Extinction Coefficient
2.7. SDS-PAGE Analysis of Phage Structural Proteins
2.8. Transmission Electron Microscopy (TEM) Analyses
2.9. Determination of Phage Particles Physical Parameters
2.10. Host Range of Isolated Phage Particles: Spot Test and Efficiency of Plating (EOP)
2.11. One-Step Growth Curves (OSGC)
2.12. Adsorption Rate
2.13. In Vitro Phage-Bacteria Inactivation Assays
2.14. Ex Vivo Phage Treatment Experiments in Artificially Contaminated Uterus
2.15. Purification of Phage DNA and Whole Genome Sequencing
2.16. Phage Genome Assembly, Taxonomic Evaluation, Annotation, and Comparisons
2.17. Formulation of the Vaginal Eggs Integrating the Phage Cocktail
2.18. Evaluation of the Maintenance of Lytic Activity of the Phage Particles Integrated in the Vaginal Eggs
2.19. Physicochemical Characterization of the Vaginal Eggs Integrating the Phage Cocktail
2.20. Evaluation of the Cytotoxicity Potential of the Vaginal Eggs Integrating Both Phage Particles, via the Agar Disk-Diffusion Assay
2.21. Statistical Analyses
3. Results
3.1. One-Step Growth Curve Analysis

3.2. Adsorption Curve

3.3. In Vitro Phage Assays

3.4. Ex Vivo Phage Treatment Experiments in Artificially Contaminated Canine Uterus

3.5. Phage Plaque Isolation

3.6. Isolation and Characterization of Bacterial Strains Responsible for Animal Pyometra Infections
3.7. UV-Vis Spectral Scans for Determination of the Phage Particles Molar Extinction Coefficients
3.8. SDS-PAGE Analysis of Phage Structural Proteins
3.9. Transmission Electron Microscopy (TEM) Analyses
3.10. Determination of Phage Particles Physical Parameters
3.11. Host Range of the Two Newly Isolated Phages: Spot Test and Efficiency of Plating (EOP)
3.12. Genomic Characterization of Phages vB_EcoM_Uniso11 and vB_EcoM_Uniso21
3.13. Evaluation of the Maintenance of Lytic Activity of the Phage Particles Integrated in the Vaginal Eggs
3.14. Determination of the Elemental Profile of the Vaginal Egg Formulations, Devoid of Phages and Integrating Both Phage Particles, by X-ray Fluorescence with Energy Dispersion (EDXRF)
3.15. Thermal Characterization of the Vaginal Egg Formulations, Devoid of Phages and Integrating Both Phage Particles, via Differential Scanning Calorimetry (DSC)
3.16. Mechanical Analyses of the Vaginal Egg Formulations, Devoid of Phages and Integrating Both Phage Particles
3.17. Surface Morphology of the Vaginal Egg Formulation Integrating Both Phage Particles via Scanning Electron Microscopy (SEM) Analyses
3.18. Evaluation of the Cytotoxicity Potential of the Vaginal Eggs Integrating Both Phage Particles, via the Agar Disk-Diffusion Assay
3.19. Evaluation of the Storage Stability of the Vaginal Eggs Integrating Both Phage Particles, in Terms of Evolution of Lytic Bioactivity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef]
- Pessanha, R.P.; Gontijo Filho, P.P. Antimicrobial use as growth promoters and resistance for isolates of Escherichia coli and of Enterobacteriaceae lactose negative of the fecal microflora of broiler chickens. Arq. Bras. Med. Vet. Zootec. 2001, 53, 111–115. [Google Scholar] [CrossRef]
- Borie, C.; Robeson, J.; Galarce, N. Lytic bacteriophages in Veterinary Medicine: A therapeutic option against bacterial pathogens? Arch. Med. Vet. 2014, 46, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Alomari, M.M.M.; Dec, M.; Urban-Chmiel, R. Bacteriophages as an Alternative Method for Control of Zoonotic and Foodborne Pathogens. Viruses 2021, 13, 2348. [Google Scholar] [CrossRef]
- Wronski, J.G.; Argenta, F.F.; Raiter, J.; Ehlers, L.P.; Sala, R.D.V.; Siqueira, F.M.; Cardoso, D.F.; Sonne, L.; Pavarini, S.P. Bilateral pyelonephritis due to Escherichia coli infection in a captive jaguar (Panthera onca). Brazilian J. Vet. Res. 2020, 40, 554–558. [Google Scholar] [CrossRef]
- Zachary, J.F. Mechanisms of microbial infections. In Pathologic Basis of Veterinary Disease; Zachary, J.F., Ed.; Elsevier: St. Louis, MI, USA, 2017; pp. 132–241. ISBN 9780323357753. [Google Scholar]
- Bazzaz, B.S.F.; Fork, S.D.; Ahmadi, R.; Khameneh, B. Deep insights into urinary tract infections and effective natural remedies. Afr. J. Urol. 2021, 27, 6. [Google Scholar] [CrossRef]
- Camargo, L.R.P.; Suffredini, I.B. Anti-Escherichia coli activity of Brazilian plant extracts: New trends in veterinary research. Arq. Bras. Med. Veterinária Zootec. 2014, 66, 617–620. [Google Scholar] [CrossRef] [Green Version]
- Lavon, Y.; Leitner, G.; Kressel, Y.; Ezra, E.; Wolfenson, D. Comparing effects of bovine Streptococcus and Escherichia coli mastitis on impaired reproductive performance. J. Dairy Sci. 2019, 102, 10587–10598. [Google Scholar] [CrossRef]
- Machado, V.S.; Bicalho, R.C. The infectious disease epidemiologic triangle of bovine uterine diseases. Anim. Reprod. 2015, 12, 450–464. [Google Scholar]
- Lima, F.S. Recent advances and future directions for uterine diseases diagnosis, pathogenesis, and management in dairy cows. Anim. Reprod. 2020, 17, e20200063. [Google Scholar] [CrossRef]
- Gonzalez Moreno, C.; Torres Luque, A.; Oliszewski, R.; Rosa, R.J.; Otero, M.C. Characterization of native Escherichia coli populations from bovine vagina of healthy heifers and cows with postpartum uterine disease. PLoS ONE 2020, 15, e0228294. [Google Scholar] [CrossRef]
- Jeon, S.J.; Galvão, K.N. An advanced understanding of uterine microbial ecology associated with metritis in dairy cows. Genom. Inform. 2018, 16, e21. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, C.F.P.M.; Borba, H.R.; Ribas, J.A.S.; Barros, R.R. Bacteriological, cytological and histopathological diagnosis of equine endometritis. Rev. Bras. Ciência Veterinária 2011, 18, 19–22. [Google Scholar] [CrossRef]
- Christoffersen, M.; Woodward, E.; Bojesen, A.M.; Jacobsen, S.; Petersen, M.R.; Troedsson, M.H.T.; Lehn-Jensen, H. Inflammatory responses to induced infectious endometritis in mares resistant or susceptible to persistent endometritis. BMC Vet. Res. 2012, 8, 41. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Dhaliwal, G.S. Escherichia coli lipopolysaccharide-induced immunomodulation along with oxytocin administration after mating as a treatment protocol for persistent endometritis in mares. J. Equine Vet. Sci. 2010, 30, 259–265. [Google Scholar] [CrossRef]
- El-Shalofy, A.S.; Derbala, M.K.; Asfour, H.A.; Eissa, H.M.; Aly, A.B. Infectious endometritis in Arabian mares: An updated clinical investigation of uterine microbial isolates, antimicrobial sensitivities and fertility in Egypt. Thai. J. Vet. Med. 2021, 51, 177–184. [Google Scholar] [CrossRef]
- Sales, K.K.S.; Rodrigues, N.M.; Rufino, A.K.B.; Luz, P.M.S. Pyometra and vaginal hyperplasia in a female dog: Case report. Pubvet 2017, 11, 78–81. [Google Scholar] [CrossRef] [Green Version]
- Sant’Anna, M.C.; Giordano, L.G.P.; Flaiban, K.K.M.C.; Muller, E.E.; Martins, M.I.M. Prognostic markers of canine pyometra. Arq. Bras. Med. Vet. Zootec. 2014, 66, 1711–1717. [Google Scholar] [CrossRef] [Green Version]
- Hagman, R. Pyometra in small animals. Vet. Clin. Small Anim. Pract. 2018, 48, 639–661. [Google Scholar] [CrossRef]
- Raheel, I.A.E.R.; Hassan, W.H.; Salem, S.S.R.; Salam, H.S.H. Biofilm forming potentiality of Escherichia coli isolated from bovine endometritis and their antibiotic resistance profiles. J. Adv. Vet. Anim. Res. 2020, 7, 442–451. [Google Scholar] [CrossRef]
- Lara, V.M.; Donadeli, M.P.; Cruz, F.S.F.; Carregaro, A.B. Antimicrobial multi-resistance of Escherichia coli strains isolated from female dogs with pyometra. Arq. Bras. Med. Vet. Zootec. 2008, 60, 1032–1034. [Google Scholar] [CrossRef]
- Dyba, S.; Oliveira, C.R.T.; Hadi, N.I.I.A.; Moutinho, I.; Oliveira, V.M.; Oliveira, V.C.; Gonçalves, G.F.; Elias, F.; Dalmolin, F. Cystic endometrial hyperplasy—Piometra in female dogs: Retrospective study and microbiological evaluation at Paraná southwest. Brazilian J. Anim. Environ. Res. 2021, 4, 1653–1666. [Google Scholar] [CrossRef]
- Coggan, J.A.; Melville, P.A.; Oliveira, C.M.; Faustino, M.; Moreno, A.M.; Benites, N.R. Microbiological and histopathological aspects of canine pyometra. Brazilian J. Microbiol. 2008, 39, 477–483. [Google Scholar] [CrossRef] [Green Version]
- Agostinho, J.M.A.; Souza, A.; Schocken-Iturrino, R.P.; Beraldo, L.G.; Borges, C.A.; Ávila, F.A.; Marin, J.M. Escherichia coli strains isolated from the uteri horn, mouth, and rectum of bitches suffering from pyometra: Virulence factors, antimicrobial susceptibilities, and clonal relationships among strains. Int. J. Microbiol. 2014, 2014, 979584. [Google Scholar] [CrossRef] [Green Version]
- Anjos, M.S.; Bittencourt, R.F.; Biscarde, C.E.A.; Silva, M.A.A.; Santos, E.S.; Maggitti Junior, L.D.P.; Santana, L.R.; Felix, M.D.; Bittencourt, M.V.; Cavalcante, A.K.S. Canine pyometra: Interferences of age and type in blood count and serum biochemistry. R. Bras. Ci. Vet. 2021, 28, 167–173. [Google Scholar] [CrossRef]
- Oliveira, F.S.; Paz, L.N.; Mota, T.M.; Oriá, A.P.; Da Silva, M.C.A.; Pinna, M.H. Profile of resistance of Escherichia coli isolated from canine pyometra. Cienc. Anim. Bras. (Goiânia) 2016, 17, 615–621. [Google Scholar] [CrossRef]
- Rocha, R.A.; Ribeiro, W.M.; Almeida, J.A.; Santos, A.L.; Fernandes, M.R.; Barbosa, M.S.; Moraes Filho, A.V.; Carneiro, L.C.; Silva, C.A. Detection of resistance genes in pyometra isolated bacteria in bitches. Brazilian J. Vet. Res. Anim. Sci. 2021, 58, e173908. [Google Scholar] [CrossRef]
- Keid, L.B.; Chiebao, D.P.; Batinga, M.C.A.; Faita, T.; Diniz, J.A.; Oliveira, T.M.F.S.; Ferreira, H.L.; Soares, R.M. Brucella canis infection in dogs from commercial breeding kennels in Brazil. Transbound Emerg Dis. 2017, 64, 691–697. [Google Scholar] [CrossRef]
- Evangelista, L.S.M.; Quessada, A.M.; Lopes, R.R.F.B.; Alves, R.P.A.; Gonçalves, L.M.F.; Drumond, K.O. Clinic and laboratory profile of cats with pyometra before and after ovaryhysterectomy. Rev. Bras. Reprod. Anim. (Belo Horizonte) 2011, 35, 347–351. [Google Scholar]
- Bezerra, W.G.A.; Horn, R.H.; Silva, I.N.G.; Teixeira, R.S.C.; Lopes, E.S.; Albuquerque, Á.H.; Cardoso, W.C. Antibiotics in the poultry industry—A review on antimicrobial resistance. Arch. Zootec. 2017, 66, 301–307. [Google Scholar]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef]
- Haq, I.U.; Chaudhry, W.N.; Akhtar, M.N.; Andleeb, S.; Qadri, I. Bacteriophages and their implications on future biotechnology: A review. Virol. J. 2012, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Rios, A.C.; Moutinho, C.G.; Pinto, F.C.; Del Fiol, F.S.; Jozala, A.F.; Chaud, M.V.; Vila, M.M.D.C.; Teixeira, J.A.; Balcão, V.M. Alternatives to overcoming bacterial resistances: State-of-the-art. Microbiol. Res. 2016, 191, 51–80. [Google Scholar] [CrossRef]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.M.D.C.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological applications of bacteriophages: State of the art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef]
- Harada, L.K.; Silva, E.C.; Rossi, F.P.N.; Cieza, B.; Oliveira, T.J.; Pereira, C.; Tomazetto, G.; Silva, B.B.; Squina, F.M.; Vila, M.M.D.C.; et al. Characterization and in vitro testing of newly isolated lytic bacteriophages for biocontrol of Pseudomonas aeruginosa. Future Microbiol. 2022, 17, 111–141. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [Green Version]
- Górski, A.; Międzybrodzki, R.; Węgrzyn, G.; Jończyk-Matysiak, E.; Borysowski, J.; Weber-Dąbrowska, B. Phage therapy: Current status and perspectives. Med. Res. Rev. 2020, 40, 459–463. [Google Scholar] [CrossRef]
- Guo, M.; Gao, Y.; Xue, Y.; Liu, Y.; Zeng, X.; Cheng, Y.; Ma, J.; Wang, H.; Sun, J.; Wang, Z.; et al. Bacteriophage Cocktails Protect Dairy Cows Against Mastitis Caused By Drug Resistant Escherichia coli Infection. Front. Cell. Infect. Microbiol. 2021, 11, 690377. [Google Scholar] [CrossRef]
- Loh, B.; Gondil, V.S.; Manohar, P.; Khan, F.M.; Yang, H.; Leptihn, S. Encapsulation and delivery of therapeutic phages. Appl. Environ. Microbiol. 2020, 87, e01979-20. [Google Scholar] [CrossRef]
- Rahimzadeh, G.; Saeedi, M.; Moosazadeh, M.; Hashemi, S.M.H.; Babaei, A.; Rezai, M.S.; Kamel, K.; Asare-Addo, K.; Nokhodchi, A. Encapsulation of bacteriophage cocktail into chitosan for the treatment of bacterial diarrhea. Sci. Rep. 2021, 11, 15603. [Google Scholar] [CrossRef]
- Malik, D.J.; Sokolov, I.J.; Vinner, G.K.; Mancuso, F.; Cinquerrui, S.; Vladisavljevic, G.T.; Clokie, M.R.J.; Garton, N.J.; Stapley, A.G.F.; Kirpichnikova, A. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv. Colloid Interface Sci. 2017, 249, 100–133. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Li, J.; Huang, H.; Wang, Y.; Qian, X.; Ren, J.; Xue, F.; Dai, J.; Tang, F. Microencapsulated phages show prolonged stability in gastrointestinal environments and high therapeutic efficiency to treat Escherichia coli O157:H7 infection. Vet. Res. 2021, 52, 118. [Google Scholar] [CrossRef]
- Gondil, V.S.; Chhibber, S. Bacteriophage and endolysin encapsulation systems: A promising strategy to improve therapeutic outcomes. Front. Pharmacol. 2021, 12, 675440. [Google Scholar] [CrossRef]
- Rosner, D.; Clark, J. Formulations for Bacteriophage Therapy and the Potential Uses of Immobilization. Pharmaceuticals 2021, 14, 359. [Google Scholar] [CrossRef]
- Balcão, V.M.; Moreli, F.C.; Silva, E.C.; Belline, B.G.; Martins, L.F.; Rossi, F.P.N.; Pereira, C.; Vila, M.M.D.C.; da Silva, A.M. Isolation and Molecular Characterization of a Novel Lytic Bacteriophage That Inactivates MDR Klebsiella pneumoniae Strains. Pharmaceutics 2022, 14, 1421. [Google Scholar] [CrossRef]
- Silva, E.C.; Oliveira, T.J.; Moreli, F.C.; Harada, L.K.; Vila, M.M.D.C.; Balcão, V.M. Newly isolated lytic bacteriophages for Staphylococcus intermedius, structurally and functionally stabilized in a hydroxyethylcellulose gel containing choline geranate: Potential for transdermal permeation in veterinary phage therapy. Res. Vet. Sci. 2021, 135, 42–58. [Google Scholar] [CrossRef]
- Adams, M.H. Bacteriophages; Interscience Publishers Inc.: New York, NY, USA; London, UK, 1959. [Google Scholar]
- Rios, A.C.; Vila, M.M.D.C.; Lima, R.; Del Fiol, F.S.; Tubino, M.; Teixeira, J.A.; Balcão, V.M. Structural and functional stabilization of bacteriophage particles within the aqueous core of a W/O/W multiple emulsion: A potential biotherapeutic system for the inhalational treatment of bacterial pneumonia. Process Biochem. 2018, 64, 177–192. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, L.A.M.; Pereira, C.; Frazão, C.; Balcão, V.M.; Almeida, A. Efficiency of Phage φ6 for Biocontrol of Pseudomonas syringae pv. syringae: An in Vitro Preliminary Study. Microorganisms 2019, 7, 286. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, L.A.M.; Pereira, C.; Barreal, M.E.; Gallego, P.P.; Balcão, V.M.; Almeida, A. Use of phage ϕ6 to inactivate Pseudomonas syringae pv. actinidiae in kiwifruit plants: In vitro and ex vivo experiments. Appl. Microbiol. Biotechnol. 2019, 104, 1319–1330. [Google Scholar] [CrossRef]
- Melo, L.D.R.; Sillankorva, S.; Ackermann, H.-W.; Kropinski, A.M.; Azeredo, J.; Cerca, N. Isolation and characterization of a new Staphylococcus epidermidis broad-spectrum bacteriophage. J. Gen. Virol. 2014, 95, 506–515. [Google Scholar] [CrossRef]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [Green Version]
- Stuer-Lauridsen, B.; Janzen, T.; Schnabl, J.; Johansen, E. Identification of the host determinant of two prolate-headed phages infecting Lactococcus lactis. Virology 2003, 309, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, K.; Takesue, S.; Ishibashi, K.; Nakahara, S. A computer simulation of the adsorption of Lactobacillus phage PL-1 to host cells: Some factors affecting the process. Agric. Biol. Chem. 1982, 46, 697–702. [Google Scholar] [CrossRef]
- Storms, Z.J.; Sauvageau, D. Modeling tailed bacteriophage adsorption: Insight into mechanisms. Virology 2015, 485, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Kokot, M.; Dlugosz, M.; Deorowicz, S. KMC 3: Counting and manipulating k-mer statistics. Bioinformatics 2017, 33, 2759–2761. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Florea, L.; Langmead, B. Lighter: Fast and memory-efficient sequencing error correction without counting. Genome Biol. 2014, 15, 509. [Google Scholar] [CrossRef] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, Y.; Watai, H.; Honda, T.; Mihara, T.; Omae, K.; Roux, S.; Blanc-Mathieu, R.; Yamamoto, K.; Hingamp, P.; Sako, Y.; et al. Environmental Viral Genomes Shed New Light on Virus-Host Interactions in the Ocean. mSphere 2017, 2, e00359-16. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 1, 421. [Google Scholar] [CrossRef] [Green Version]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [Green Version]
- Cabanettes, F.; Klopp, C. D-GENIES: Dot plot large genomes in an interactive, efficient and simple way. PeerJ. 2018, 6, e4958. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. PILER-CR: Fast and accurate identification of CRISPR repeats. BMC Bioinform. 2007, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchfink, B.; Reuter, K.; Drost, H.G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [Green Version]
- Sayers, E.W.; Agarwala, R.; Bolton, E.E.; Brister, J.R.; Canese, K.; Clark, K.; Connor, R.; Fiorini, N.; Funk, K.; Hefferon, T.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019, 47, D23–D28. [Google Scholar] [CrossRef] [Green Version]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bairoch, A. UniProtKB/Swiss-Prot. Methods Mol. Biol. 2007, 406, 89–112. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [Green Version]
- Pedruzzi, I.; Rivoire, C.; Auchincloss, A.H.; Coudert, E.; Keller, G.; de Castro, E.; Baratin, D.; Cuche, B.A.; Bougueleret, L.; Poux, S.; et al. HAMAP in 2015: Updates to the protein family classification and annotation system. Nucleic Acids Res. 2015, 43, D1064–D1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Boscariol, R.; Oliveira Junior, J.M.; Baldo, D.A.; Balcão, V.M.; Vila, M.M.D.C. Transdermal permeation of curcumin promoted by choline geranate ionic liquid: Potential for the treatment of skin diseases. Saudi Pharm. J. 2022, 30, 382–397. [Google Scholar] [CrossRef]
- Rocha, L.K.H.; Favaro, L.I.L.; Rios, A.C.; Silva, E.C.; Silva, W.F.; Stigliani, T.P.; Guilger, M.; Lima, R.; Oliveira, J.M.; Aranha, N.; et al. Sericin from Bombyx mori cocoons. Part I: Extraction and physicochemical-biological characterization for biopharmaceutical applications. Process Biochem. 2017, 61, 163–177. [Google Scholar] [CrossRef]
- Rogero, S.O.; Lugão, A.B.; Ikeda, T.I.; Cruz, A.S. In vitro cytotoxicity test: Comparative study between two methodologies. Mat. Res. 2003, 6, 317–320. [Google Scholar] [CrossRef]
- Pusnik, M.; Imeri, M.; Deppierraz, G.; Bruinink, A.; Zinn, M. The agar diffusion scratch assay—A novel method to assess the bioactive and cytotoxic potential of new materials and compounds. Sci. Rep. 2016, 6, 20854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorge, L.R.; Harada, L.K.; Silva, E.C.; Campos, W.F.; Oliveira, J.M.; Vila, M.M.D.C.; Tubino, M.; Balcão, V.M. Bacterial nanocellulose biomembrane as a support for human insulin aiming at transdermal permeation. Quim. Nova 2020, 43, 572–578. [Google Scholar] [CrossRef]
- Bates, D.M.; Watts, D.G. Nonlinear Regression Analysis and Its Applications, 1st ed.; Wiley-Blackwell: Hoboken, NJ, USA, 1988; ISBN 978-0471816430. [Google Scholar]
- Balcão, V.M.; Vieira, M.C.; Malcata, F.X. Adsorption of Protein from Several Commercial Lipase Preparations onto a Hollow-Fiber Membrane Module. Biotechnol. Prog. 1996, 12, 164–172. [Google Scholar] [CrossRef]
- Balcão, V.M.; Oliveira, T.A.; Xavier Malcata, F. Stability of a commercial lipase from Mucor javanicus: Kinetic modelling of pH and temperature dependencies. Biocatal. Biotransform. 1998, 16, 45–66. [Google Scholar] [CrossRef]
- Gómez-Gómez, J.-M.; Manfredi, C.; Alonso, J.-C.; Blázquez, J. A novel role for RecA under non-stress: Promotion of swarming motility in Escherichia coli K-12. BMC Biol. 2007, 5, 14. [Google Scholar] [CrossRef] [Green Version]
- Pires, D.P.; Oliveira, H.; Melo, L.D.R.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, H.; São-José, C.; Azeredo, J. Phage-derived peptidoglycan degrading enzymes: Challenges and future prospects for in vivo therapy. Viruses 2018, 10, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateus, L.; Henriques, S.; Merino, C.; Pomba, C.; Costa, L.L.; Silva, E. Virulence genotypes of Escherichia coli canine isolates from pyometra, cystitis and fecal origin. Vet. Microbiol. 2013, 166, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J. Application of UV Light Scattering to Detect Reversible Self-association and Aggregation of Proteins in Solution, University of Connecticut. Ph.D. Thesis, University of Connecticut, Storrs, CT, USA, 2017. [Google Scholar]
- Vekshin, N.L. Screening Hypochromism of Chromophores in Macromolecular Biostructures. J. Biol. Phys. 1999, 25, 339–354. [Google Scholar] [CrossRef]
- Kakasis, A.; Panitsa, G. Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int. J. Antimicrob. Agents 2019, 53, 16–21. [Google Scholar] [CrossRef]
- Hughes, K.A.; Sutherland, I.W.; Clark, J.; Jones, M.V. Bacteriophage and associated polysaccharide depolymerases—Novel tools for study of bacterial biofilms. J. Appl. Microbiol. 2002, 85, 583–590. [Google Scholar] [CrossRef]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [Green Version]
- Seed, K.D. Battling phages: How bacteria defend against viral attack. PLOS Pathog. 2015, 11, e1004847. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Cunha, Â.; Delgadillo, I.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Bacteriophages with potential to inactivate Salmonella Typhimurium: Use of single phage suspensions and phage cocktails. Virus Res. 2016, 220, 179–192. [Google Scholar] [CrossRef]
- Abedon, S.T.; Culler, R.R. Optimizing bacteriophage plaque fecundity. J. Theor. Biol. 2007, 249, 582–592. [Google Scholar] [CrossRef]
- Mateus, L.; Costa, L.; Silva, Y.J.; Pereira, C.; Cunha, A.; Almeida, A. Efficiency of phage cocktails in the inactivation of Vibrio in aquaculture. Aquaculture 2014, 424–425, 167–173. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Li, Z.J.; Wang, S.-W.; Wang, S.-M.; Huang, D.-H.; Li, Y.-H.; Ma, Y.-Y.; Wang, J.; Liu, F.; Chen, X.-D.; et al. Isolation and characterization of ZZ1, a novel lytic phage that infects Acinetobacter baumannii clinical isolates. BMC Microbiol. 2012, 12, 156. [Google Scholar] [CrossRef]
- Ceyssens, P.-J. Isolation and characterization of lytic bacteriophages infecting Pseudomonas aeruginosa, Katholieke Inuversiteit Leuven. Ph.D. Thesis, Katholieke Universiteit Leuven, Leuven, Belgium, December 2009. [Google Scholar]
- Hyman, P.; Abedon, S.T. Practical methods for determining phage growth parameters. In Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 501, pp. 175–202. ISBN 9781603271646. [Google Scholar]
- Moldovan, R.; Chapman-McQuiston, E.; Wu, X.L. On kinetics of phage adsorption. Biophys. J. 2007, 93, 303–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakhuba, D.V.; Kolomiets, E.I.; Szwajcer Dey, E.; Novik, G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Polish J. Microbiol. 2010, 59, 145–155. [Google Scholar] [CrossRef]
- Heller, K.; Braun, V. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. J. Bacteriol. 1979, 139, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Zemb, O.; Manefield, M.; Thomas, F.; Jacquet, S. Phage adsorption to bacteria in the light of the electrostatics: A case study using E. coli, T2 and flow cytometry. J. Virol. Methods 2013, 189, 283–289. [Google Scholar] [CrossRef]
- Nabergoj, D.; Modic, P.; Podgornik, A. Effect of bacterial growth rate on bacteriophage population growth rate. MicrobiologyOpen 2018, 7, e558. [Google Scholar] [CrossRef] [Green Version]
- Tsukada, K.; Okazaki, M.; Kita, H.; Inokuchi, Y.; Urabe, I.; Yomo, T. Quantitative analysis of the bacteriophage Qβ infection cycle. Biochim. Biophys. Acta 2009, 1790, 65–70. [Google Scholar] [CrossRef]
- Høyland-Kroghsbo, N.M.; Mærkedahl, R.B.; Svenningsen, S.L. A quorum-sensing-induced bacteriophage defense mechanism. mBio 2013, 4, e00362-12. [Google Scholar] [CrossRef] [Green Version]
- Balcão, V.M.; Basu, A.; Cieza, B.; Rossi, F.N.; Pereira, C.; Vila, M.M.D.C.; Setubal, J.C.; Ha, T.; da Silva, A.M. Pseudomonas-tailed lytic phages: Genome mechanical analysis and putative correlation with virion morphogenesis yield. Future Microbiol. 2022, 17, 1009–1026. [Google Scholar] [CrossRef] [PubMed]
- Shephard, J.; McQuillan, A.J.; Bremer, P.J. Mechanisms of Cation Exchange by Pseudomonas aeruginosa PAO1 and PAO1 wbpL, a Strain with a Truncated Lipopolysaccharide. Appl. Environ. Microbiol. 2008, 74, 6980–6986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteban, P.P.; Jenkins, A.T.A.; Arnot, T.C. Elucidation of the mechanisms of action of Bacteriophage K/nano-emulsion formulations against S. aureus via measurement of particle size and zeta potential. Colloids Surfaces B Biointerfaces 2016, 139, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Liao, C.; Thompson, M.L.; Soupir, M.L.; Jarboe, L.R.; Dixon, P.M. E. coli surface properties differ between stream water and sediment environments. Front. Microbiol. 2016, 7, 1732. [Google Scholar] [CrossRef] [Green Version]
- Hamadi, F.; Latrache, H.; Zahir, H.; Elghmari, A.; Timinouni, M.; Ellouali, M. The relation between Escherichia coli surface functional groups’ composition and their physicochemical properties. Brazilian J. Microbiol. 2008, 39, 10–15. [Google Scholar] [CrossRef] [Green Version]
- Glasser, C.A.; Vila, M.M.D.C.; Pereira, J.C.; Chaud, M.V.; Oliveira Júnior, J.M.; Tubino, M.; Balcão, V.M. Development of a water-in-oil-in-water multiple emulsion system integrating biomimetic aqueous-core lipid nanodroplets for protein entity stabilization. Part II: Process and product characterization. Drug Dev. Ind. Pharm. 2016, 42, 1990–2000. [Google Scholar] [CrossRef]
- Katayama, K.; Nomura, H.; Ogata, H.; Eitoku, T. Diffusion coefficients for nanoparticles under flow and stop-flow conditions. Phys. Chem. Chem. Phys. 2009, 11, 10494–10499. [Google Scholar] [CrossRef]
- Chi-Hsin, H.; ChongYi, L.; JongKang, L.; Chan-Shing, L. Control of the eel (Anguilla japonica) pathogens, Aeromonas hydrophila and Edwardsiella tarda, by bacteriophages. J. Fish. Soc. Taiwan 2000, 27, 21–31. [Google Scholar]
- Pasharawipas, T.; Manopvisetcharean, J.; Flegel, T.W. Phage treatment of Vibrio harveyi: A general concept of protection against bacterial infection. Res. J. Microbiol. 2011, 6, 560–567. [Google Scholar] [CrossRef] [Green Version]
- Prasad, Y.; Arpana; Kumar, D.; Sharma, A.K. Lytic bacteriophages specific to Flavobacterium columnare rescue catfish, Clarias batrachus (Linn.) from columnaris disease. J. Environ. Biol. 2011, 32, 161–168. [Google Scholar]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Characterization and in vitro evaluation of new bacteriophages for the biocontrol of Escherichia coli. Virus Res. 2017, 227, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.; Pereira, C.; Moreirinha, C.; Salvio, R.; Lopes, A.; Wang, D.; Almeida, A. New insights on phage efficacy to control Aeromonas salmonicida in aquaculture systems: An in vitro preliminary study. Aquaculture 2018, 495, 970–982. [Google Scholar] [CrossRef]
- Costa, P.; Pereira, C.; Gomes, A.T.P.C.; Almeida, A. Efficiency of single phage suspensions and phage cocktail in the inactivation of Escherichia coli and Salmonella Typhimurium: An in vitro preliminary study. Microorganisms 2019, 7, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forti, F.; Roach, D.R.; Cafora, M.; Pasini, M.E.; Horner, D.S.; Fiscarelli, E.V.; Rossitto, M.; Cariani, L.; Briani, F.; Debarbieux, L.; et al. Design of a Broad-Range Bacteriophage Cocktail That Reduces Pseudomonas aeruginosa Biofilms and Treats Acute Infections in Two Animal Models. Antimicrob. Agents Chemother. 2018, 62, e02573-17. [Google Scholar] [CrossRef] [Green Version]
- Gill, J.J.; Hyman, P. Phage choice, isolation, and preparation for phage therapy. Curr. Pharm. Biotechnol. 2010, 11, 2–14. [Google Scholar] [CrossRef]
- Campos, W.F.; Silva, E.C.; Oliveira, T.J.; Oliveira, J.M., Jr.; Tubino, M.; Pereira, C.; Vila, M.M.D.C.; Balcão, V.M. Transdermal permeation of bacteriophage particles by choline oleate: Potential for treatment of soft-tissue infections. Future Microbiol. 2020, 15, 881–896. [Google Scholar] [CrossRef]
- Harada, L.K.; Bonventi Júnior, W.; Silva, E.C.; Oliveira, T.J.; Moreli, F.C.; Oliveira Júnior, J.M.; Tubino, M.; Vila, M.M.D.C.; Balcão, V.M. Bacteriophage-based biosensing of Pseudomonas aeruginosa: An integrated approach for the putative real-time detection of multi-drug-resistant strains. Biosensors 2021, 11, 124. [Google Scholar] [CrossRef]
- Chen, W.; Duša, F.; Witos, J.; Ruokonen, S.-K.; Wiedmer, S.K. Determination of the main phase transition temperature of phospholipids by nanoplasmonic sensing. Sci. Rep. 2018, 8, 14815. [Google Scholar] [CrossRef] [Green Version]
- Lewis, R.N.A.H.; Zhang, Y.-P.; McElhaney, R.N. Calorimetric and spectroscopic studies of the phase behavior and organization of lipid bilayer model membranes composed of binary mixtures of dimyristoylphosphatidylcholine and dimyristoylphosphatidylglycerol. Biochim. Biophys. Acta-Biomembr. 2005, 1668, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Segman, O.; Wiesman, Z.; Yarmolinsky, L. Methods and technologies related to shea butter chemophysical properties and to the delivery of bioactives in chocolate and related products. In Cocoa Butter and Related Compounds; Garti, N., Widlak, N.R., Eds.; AOCS Press: Urbana, IL, USA, 2012; pp. 417–441. ISBN 9780983079125. [Google Scholar]
- Minifie, B.W. Cocoa Butter and Replacement Fats. In Chocolate, Cocoa and Confectionery: Science and Technology; Springer: Dordrecht, The Netherlands, 1989; ISBN 978-94-011-7926-3. [Google Scholar]
- Kishino, T.; Inoue, Y.; Inoue, K.; Saito, K.; Kondo, M.; Otsuka, K.; Terukina, T.; Hattori, Y.; Otsuka, M. Pharmaceutical evaluation of hospital-prepared, ulinastatin-containing vaginal suppositories. J. Drug Deliv. Sci. Technol. 2018, 44, 1–7. [Google Scholar] [CrossRef]
- Kishino, T.; Inoue, Y.; Inoue, K.; Saito, K.; Kondo, M.; Otsuka, K.; Terukina, T.; Hattori, Y.; Otsuka, M. Predictive Evaluation of Pharmaceutical Properties of Ulinastatin-Containing Vaginal Suppositories as a Hospital Preparation by Near-Infrared Spectroscopy. Chem. Pharm. Bull. 2018, 66, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceschel, G.C.; Maffei, P.; Lombardi Borgia, S.; Ronchi, C.; Rossi, S. Development of a mucoadhesive dosage form for vaginal administration. Drug Dev. Ind. Pharm. 2001, 27, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Pashayan, M.M.; Hovhannisyan, H.G. Development of bifunctional vaginal suppositories by joint use terconazole and probiotic for treatment and prophylaxis of vulvovaginal candidiasis. Drug Dev. Ind. Pharm. 2021, 47, 1424–1431. [Google Scholar] [CrossRef] [PubMed]















| Bacterial Strains | Source | Phage vB_EcoM_Uniso11 | Phage vB_EcoM_Uniso21 | ||
|---|---|---|---|---|---|
| Spot Test | EOP (%) | Spot Test | EOP (%) | ||
| E. coli CCCD—E003 | Collection, CEFAR | + | 100 (host) | + | 100 (host) |
| Salmonella enterica CCCD—S004 | Collection, CEFAR | - | - | - | - |
| Pseudomonas aeruginosa CCCD—P004 | Collection, CEFAR | - | - | - | - |
| Proteus mirabilis CCCD—P001 | Collection, CEFAR | - | - | - | - |
| Enterococus faecalis CCCD—E002 | Collection, CEFAR | - | - | - | - |
| Bacillus subtilis CCCD—B010 | Collection, CEFAR | - | - | - | - |
| Staphylococcus epidermis CCCD—S010 | Collection, CEFAR | - | - | - | - |
| Staphylococcus aureus CCCD—S009 | Collection, CEFAR | - | - | - | - |
| Klebsiella pneumoniae CCCD—K001 | Collection, CEFAR | - | - | - | - |
| E. coli ATCC-25922 | Collection, ATCC | + | 44.33 | + | 37.66 |
| Enterobacter sp. (*2.2) | Collection, Labiton-UNISO | - | - | - | - |
| Enterobacter aerogenes (*2.13) | Collection, Labiton-UNISO | - | - | - | - |
| Klebsiella pneumoniae (*4.15) | Collection, Labiton-UNISO | - | - | - | - |
| Staphylococcus intermedius* | Collection, PhageLab-UNISO | - | - | - | - |
| Pseudomonas syringae pv. syringae* | Collection, PhageLab-UNISO | - | - | - | - |
| Pseudomonas syringae pv. garcae* | Collection, PhageLab-UNISO | - | - | - | - |
| Xanthomonas axonopodis pv. citri 306 | Collection, PhageLab-UNISO | - | - | - | - |
| Proteus penneri (*5.5) | Collection, Labiton-UNISO | - | - | - | - |
| Proteus vulgaris (*5.4) | Collection, Labiton-UNISO | - | - | - | - |
| E. coli (*3.2) | Collection, Labiton-UNISO | + | 12.89 | + | 9.54 |
| E. coli (*3.4) | Collection, Labiton-UNISO | + | 45.66 | + | 41.34 |
| E. coli (*3.5) | Collection, Labiton-UNISO | - | - | - | - |
| E. coli (*A1) | Clinical isolate, non-castrated female cat | + | 55.66 | + | 59.36 |
| E. coli (*A4) | Clinical isolate, non-castrated female cat | + | 48.21 | + | 39.18 |
| E. coli (*A5) | Clinical isolate, female dog with pyometra | + | 10.89 | + | 12.18 |
| E. coli (*A6) | Clinical isolate, female dog with pyometra | - | - | - | - |
| E. coli (*A7) | Clinical isolate, female dog with pyometra | + | 26.81 | + | 32.64 |
| E. coli (*A8) | Clinical isolate, female dog with pyometra | + | 70.66 | + | 71.12 |
| E. coli (*A9) | Clinical isolate, female dog with pyometra | + | 29.33 | + | 19.54 |
| E. coli (*A10) | Clinical isolate, female dog with pyometra | - | - | - | - |
| E. coli (*A11) | Clinical isolate, female dog with pyometra | + | 45.34 | + | 52.33 |
| E. coli (*A14) | Clinical isolate, female dog with pyometra | + | 88.23 | + | 76.54 |
| E. coli (*A16) | Clinical isolate, female dog with pyometra | - | - | - | - |
| E. coli (*A17) | Clinical isolate, non-castrated female cat | + | 59.66 | + | 44.18 |
| E. coli (*A19) | Clinical isolate, non-castrated female cat | + | 64.54 | + | 70.12 |
| E. coli (*A21, #1) | Clinical isolate, mare | + | 0.00016 | + | 0.0000005 |
| E. coli (*A22, #2) | Clinical isolate, mare | - | - | - | - |
| E. coli (*A24, #4) | Clinical isolate, mare | - | - | - | - |
| E. coli (*A25, #5) | Clinical isolate, mare | - | - | - | - |
| E. coli (*A26, #6) | Clinical isolate, mare | + | 0.00014 | + | 0.000002 |
| E. coli (*A27, #7) | Clinical isolate, mare | - | - | - | - |
| Components | Function in the Formulation | Amount (mg) | % (w/w) | |
|---|---|---|---|---|
| Plain Vaginal Eggs | Antibacterial Vaginal Eggs | |||
| Cocoa butter | Lipid matrix | 9842 | 9842 | 98.42 |
| Concentrated phage VB_EcoM_US_11 suspension (2.38 × 1011 PFU/mL) | Active antibacterial entities | 0 | 9 (9 µL, containing 2.142 × 109 virions) | 0.180 |
| Concentrated phage VB_EcoM_US_21 suspension (2.12 × 1011 PFU/mL) | 0 | 9 (9 µL, containing 1.908 × 109 virions) | ||
| Ultrapure water | Plasticizer | 18 | 0 | |
| Tween 80 | Nonionic (polysorbate) surfactant to help stabilize the bacteriophage particles | 40 | 40 | 0.400 |
| Preservative (methylparaben, 6 g; propylparaben, 3 g; propylenglycol, 91 g) | Preservative, antifungal | 100 | 100 | 1.000 |
| TOTAL: | 10,000 | 10,000 | 100.00 | |
| Clinical Sample | Origin | Gram Coloration | Lactose Fermentation | Oxidase | Citrate | Gas | Indol | Lysin | Motility | H2S Production | Urea | Identification |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | Cat | - | + | - | - | + | + | + | - | - | - | E. coli |
| A2 | Canine | + | n.p. | n.p. | n.p. | n.p. | n.p. | n.p. | n.p. | n.p. | n.p. | Streptococcus sp. |
| A3 | Canine | - | + | - | + | + | - | + | - | - | + | Klebsiella pneumoniae |
| A4 | Cat | - | + | - | - | + | + | - | - | - | - | E. coli |
| A5 | Canine | - | + | - | - | + | + | + | + | - | - | E. coli |
| A6 | Canine | - | + | - | - | + | + | + | + | - | - | E. coli |
| A7 | Canine | - | + | - | - | + | + | + | - | - | - | E. coli |
| A8 | Canine | - | + | - | - | + | + | + | + | - | - | E. coli |
| A9 | Canine | - | + | - | - | + | + | + | - | - | - | E. coli |
| A10 | Canine | - | + | - | - | + | + | + | - | - | - | E. coli |
| A11 | Canine | - | + | - | - | + | + | + | + | - | - | E. coli |
| A12 | Canine | - | - | - | - | - | - | - | - | - | - | No growth |
| A13 | Canine | - | - | - | + | - | - | - | + | + | + | Proteus mirabilis |
| A14 | Canine | - | + | - | - | + | + | - | + | - | - | E. coli |
| A15 | Canine | - | + | - | + | + | - | + | - | - | + | Klebsiella pneumoniae |
| A16 | Canine | - | + | - | - | + | + | + | + | - | - | E. coli |
| A17 | Cat | - | + | - | - | + | + | - | + | - | - | E. coli |
| A18 | Canine | - | + | - | + | + | - | + | - | - | + | Klebsiella pneumoniae |
| A19 | Cat | - | + | - | - | + | + | + | + | - | - | E. coli |
| A20 | Canine | - | + | - | + | + | - | + | - | - | + | Klebsiella pneumoniae |
| A21 (#1) | Mare | - | + | - | - | + | + | + | - | - | - | E. coli |
| A22 (#2) | Mare | - | + | - | - | + | + | + | - | - | - | E. coli |
| A23 (#3) | Mare | - | - | - | + | + | - | + | + | - | + | Serratia marcescens |
| A24 (#4) | Mare | - | + | - | - | + | + | + | - | - | - | E. coli |
| A25 (#5) | Mare | - | + | - | - | + | + | + | - | - | - | E. coli |
| A26 (#6) | Mare | - | + | - | - | + | + | - | + | - | - | E. coli |
| A27 (#7) | Mare | - | + | - | - | + | + | - | + | - | - | E. coli |
| Physical Parameter | Phage Particles | E. coli CCCD-E003 Cells | |
|---|---|---|---|
| vB_EcoM_Uniso11 | vB_EcoM_Uniso21 | ||
| Hydrodynamic size (nm) | 310.76 ± 25.40 | 109.09 ± 3.68 | 607.52 ± 4.92 |
| Polydispersity Index | 0.383 ± 0.027 | 0.384 ± 0.023 | 0.349 ± 0.024 |
| Zeta Potential (mV) | −8.31 ± 1.12 | −16.13 ± 2.64 | −38.49 ± 0.32 |
| Electrophoretic Mobility (µ/s)/(V/cm) | −0.650 ± 0.085 | −1.260 ± 0.212 | −3.010 ± 0.030 |
| Diffusion Coefficient (m2 s−1) | 7.39 × 10−12 ± 6.27 × 10−14 | 2.25 × 10−12 ± 7.45 × 10−14 | 4.04 × 10−13 ± 3.26 × 10−15 |
| Feature | Phage vB_EcoM_Uniso11 (Phage ph0011) | Phage vB_EcoM_Uniso21 (Phage ph0021) |
|---|---|---|
| NCBI/Genbank accession number | OP557969 | OP557970 |
| Genome size | 348,288 bp | 165,222 bp |
| Number of PE reads | 23,449,832 | 20,127,644 |
| Number of PE reads mapping in the final assembly | 11,724,916 (50.0%) | 10,063,822 (50.0%) |
| Average sequencing coverage | 33.7× | 60.9× |
| GC content | 45% | 43% |
| tRNA genes | 7 + 1 pseudogene | 11 + 3 pseudogenes |
| Protein-coding genes (CDS) predicted | 575 | 264 |
| With function assigned | 128 (22.3%) | 245 (92.8%) |
| Hypothetical/unknown function | 447 (77.7%) | 19 (7.2%) |
| Closest phage genome sequence | E. coli phage vB_EcoM_phAPEC6 | E. coli phage vB_EcoM_WL-3 |
| NCBI accession number of similar phage | MK817115.1 | MT968995.1 |
| Family/Genus | Myoviridae/Asteriusvirus | Myoviridae/Tequatrovirus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balcão, V.M.; Belline, B.G.; Silva, E.C.; Almeida, P.F.F.B.; Baldo, D.Â.; Amorim, L.R.P.; Oliveira Júnior, J.M.; Vila, M.M.D.C.; Del Fiol, F.S. Isolation and Molecular Characterization of Two Novel Lytic Bacteriophages for the Biocontrol of Escherichia coli in Uterine Infections: In Vitro and Ex Vivo Preliminary Studies in Veterinary Medicine. Pharmaceutics 2022, 14, 2344. https://doi.org/10.3390/pharmaceutics14112344
Balcão VM, Belline BG, Silva EC, Almeida PFFB, Baldo DÂ, Amorim LRP, Oliveira Júnior JM, Vila MMDC, Del Fiol FS. Isolation and Molecular Characterization of Two Novel Lytic Bacteriophages for the Biocontrol of Escherichia coli in Uterine Infections: In Vitro and Ex Vivo Preliminary Studies in Veterinary Medicine. Pharmaceutics. 2022; 14(11):2344. https://doi.org/10.3390/pharmaceutics14112344
Chicago/Turabian StyleBalcão, Victor M., Bianca G. Belline, Erica C. Silva, Pablo F. F. B. Almeida, Denicezar Â. Baldo, Lara R. P. Amorim, José M. Oliveira Júnior, Marta M. D. C. Vila, and Fernando S. Del Fiol. 2022. "Isolation and Molecular Characterization of Two Novel Lytic Bacteriophages for the Biocontrol of Escherichia coli in Uterine Infections: In Vitro and Ex Vivo Preliminary Studies in Veterinary Medicine" Pharmaceutics 14, no. 11: 2344. https://doi.org/10.3390/pharmaceutics14112344