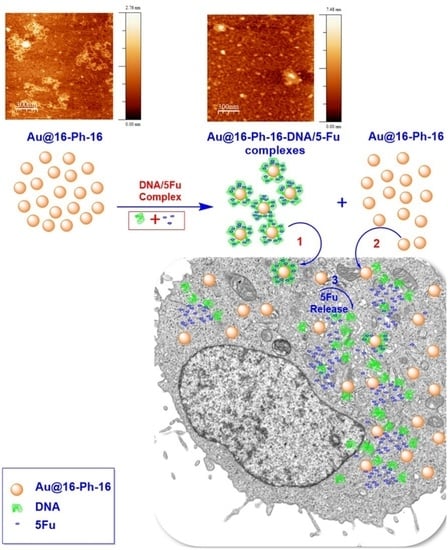
Biocompatible DNA/5-Fluorouracil-Gemini Surfactant-Functionalized Gold Nanoparticles as Promising Vectors in Lung Cancer Therapy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Synthesis of N,N’-[1,3-phenylenebis(methylene))bis[N,N’-dimethyl-N-(1-hexadecyl)]-ammonium dibromide, 16-Ph-16
2.1.2. Synthesis of Au@16-Ph-16 Precursor Gold Nanoparticles
2.1.3. Synthesis of Au@16-Ph-16/DNA–5-Fu Gold Nanoparticles
2.2. Methods
2.2.1. (3-(4,5-. dimethylthiazol-2-yl)-2,5-diphenyltetrazole) (MTT) Viability Assay
2.2.2. UV/Vis Spectroscopy
2.2.3. Fluorescence Spectroscopy
2.2.4. Circular Dichroism (CD) Spectroscopy
2.2.5. Atomic Force Microscopy Experiments
2.2.6. Dynamic Light Scattering (DLS) and Zeta Potential Measurements
2.2.7. TEM and EDS Measurements
2.2.8. Confocal Microscope
3. Results and Discussion
3.1. Exploring Experimental Conditions to Obtain Optimal Au@16-Ph-16/DNA–5-Fu Vectors. Reversible Compaction of DNA/5-Fluorouracil Complexes
3.2. Physicochemical Characterization and Stability of Au@16-Ph-16 and Au@16-Ph-16/DNA–5-Fu Nanosystems
3.3. In Vitro Biocompatibility of Gold Nanosystems
3.4. Internalization of Au@16-Ph-16 and Au@16-Ph-16/DNA–5-Fu Nanosystems
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Home Page. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer. (accessed on 18 December 2020).
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer, J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abastado, J.P.; Adams, C.; Adewole, F.; Akaza, H.; Allen, N.E.; Alsan, M.; Al Alwan, N.; Amin, M.B.; Anderson, B.O.; Anttila, A.; et al. (Eds.) World Cancer Report 2014; International Agency for Research on Cancer: Lyon, France, 2014; ISBN 978-92-832-0432-9. [Google Scholar]
- Crighton, D.; Wilkinson, S.; O’Prey, J.; Syed, N.; Smith, P.; Harrison, P.R.; Gasco, M.; Garrone, O.; Crook, T.; Ryan, K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006, 121–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- American Cancer Society (ACS). Home Page. Available online: https://www.cancer.org/ (accessed on 20 December 2020).
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasubramanian, V.; Liu, Z.; Hirvonen, J.; Santos, H.A. Bridging the knowledge of different worlds to understand the big picture of cancer nanomedicines. Adv. Healthcare Mater. 2017, 1700432, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Ramos Camos, E.V.; Rodriguez-Torres, M.P.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [Green Version]
- Ajorlou, E.; Khosroushahi, A.Y. Trends on polymer- and lipid-based nanostructures for parenteral drug delivery to tumors. Cancer Chemother. Pharmacol. 2017, 79, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Wallat, J.D.; Harrison, J.K.; Pokorski, J.K. pH responsive doxorubicin delivery by fluorous polymers for cancer treatment. Mol Pharm. 2018, 15, 2954–2962. [Google Scholar] [CrossRef] [PubMed]
- Vardhan, H.; Mittal, P.; Adena, S.K.R.; Upadhyay, M.; Yadav, S.K.; Mishra, B. Process optimization and in vivo performance of docetaxel loaded PHBV-TPGS therapeutic vesicles: A synergistic approach. Int. J. Biol. Macromol. 2018, 108, 729–743. [Google Scholar] [CrossRef]
- García-Fernández, C.; Fornaguera, C.; Borrós, S. Nanomedicine in non-small cell lung cancer: From conventional treatments to immunotherapy. Cancers 2020, 12, 1609. [Google Scholar] [CrossRef]
- Gao, J.; Ma, X.; Zhang, L.; Yan, J.; Cui, H.; Zhang, Y.; Wang, D.; Zhang, H. Self-assembled disulfide bond bearing paclitaxel-camptothecin prodrug nanoparticle for lung cancer therapy. Pharmaceutics 2020, 12, 1169. [Google Scholar] [CrossRef]
- Goudarzi, F.; Asadi, A.; Afsharpour, M.; Jamadi, R.H. In Vitro Characterization and Evaluation of the Cytotoxicity Effects of Nisin and Nisin-Loaded PLA-PEG-PLA Nanoparticles on Gastrointestinal (AGS and KYSE-30), Hepatic (HepG2) and Blood (K562) Cancer Cell Lines. AAPS Pharmscitech. 2018, 19, 1554–1566. [Google Scholar] [CrossRef] [PubMed]
- Di-Wen, S.; Pan, G.Z.; Hao, L.; Zhang, J.; Xue, Q.Z.; Wang, P.; Yuan, Q.Z. Improved antitumor activity of epirubicin-loaded CXCR4-targeted polymeric nanoparticles in liver cancers. Int. J. Pharm. 2016, 500, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Tran, P.H.; Wang, Y.; Li, P.; Kong, L. Nanoparticulate drug delivery to colorectal cancer: Formulation strategies and surface engineering. Curr. Pharm. Des. 2016, 22, 2904–2912. [Google Scholar] [CrossRef]
- Badran, M.M.; Mady, M.M.; Ghannam, M.M.; Shakeel, F. Preparation and characterization of polymeric nanoparticles surface modified with chitosan for target treatment of colorectal cancer. Int. J. Biol. Macromol. 2017, 95, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Faramarzi, L.; Dadashpour, M.; Sadeghzadeh, H.; Mahdavi, M.; Zarghami, N. Enhanced anti-proliferative and pro-apoptotic effects of metformin encapsulated PLGA-PEG nanoparticles on SKOV3 human ovarian carcinoma cells. Artif. Cells Nanomed. Biotechnol. 2019, 47, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Li, M.; Lin, Y.; Zhan, X. Encapsulation of verapamil and doxorubicin by MPEG-PLA to reverse drug resistance in ovarian cancer. Biomed. Pharmacother. 2018, 108, 565–573. [Google Scholar] [CrossRef]
- Velpurisiva, P.; Rai, P. Synergistic action of gefitinib and GSK41364A simultaneously loaded in ratiometrically-engineered polymeric nanoparticles for glioblastoma multiforme. J. Clin. Med. 2019, 8, 367. [Google Scholar] [CrossRef] [Green Version]
- Rai, M.; Ingle, A.P.; Birla, S.; Yadav, A.; Santos, C.A. Strategic role of selected noble metal nanoparticles in medicine. Crit. Rev. Microbiol. 2016, 42, 696–719. [Google Scholar] [CrossRef]
- Ghosh, P.S.; Kim, C.K.; Han, G.; Forbes, N.S.; Rotello, V.M. Efficient gene delivery vectors by tuning the surface charge density of amino acid-functionalized gold nanoparticles. ACS Nano 2008, 2, 2213–2218. [Google Scholar] [CrossRef] [Green Version]
- Ghiuță, I.; Cristea, D. Silver nanoparticles for delivery purposes. Nanoeng. Biomater. Adv. Drug Deliv. 2020, 347–371. [Google Scholar] [CrossRef]
- Tripathi, R.M.; Shrivastav, A.; Shrivastav, B.R. Biogenic gold nanoparticles: As a potential candidate for brain tumor directed drug delivery. Artif. Cell Nanomed. B. 2014, 43, 311–317. [Google Scholar] [CrossRef]
- Thorek, D.L.; Tsourkas, A. Size, charge and concentration dependent uptake of iron oxide particles by non-phagocytic cells. Biomaterials 2008, 29, 3583–3590. [Google Scholar] [CrossRef] [Green Version]
- Baranwal, A.; Mahato, K.; Srivastava, A.; Maurya, P.K.; Chandra, P. Phytofabricated metallic nanoparticles and their clinical applications. RSC Adv. 2016, 6, 105996–106010. [Google Scholar] [CrossRef]
- Mahato, K.; Nagpal, S.; Shah, M.A.; Srivastava, A.; Maurya, P.K.; Roy, S.; Jaiswal, A.; Singh, R.; Chandra, P. Gold nanoparticle surface engineering strategies and their applications in biomedicine and diagnostics. 3 Biotechnol. 2019, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Zana, R.; Benrraou, M.; Rueff, R. Alkanediyl-alpha.,.omega.-bis(dimethylalkylammonium bromide) surfactants. 1. Effect of the spacer chain length on the critical micelle concentration and micelle ionization degree. Langmuir 1991, 7, 1072. [Google Scholar] [CrossRef]
- Wettig, S.D.; Verrall, R.E.; Foldvari, M. Gemini surfactants: A new family of building blocks for non-viral gene delivery systems. Curr. Gene Ther. 2008, 8, 9–23. [Google Scholar] [CrossRef]
- He, Y.; Shang, Y.; Shao, S.; Liu, H.; Hu, Y. Micellization of cationic gemini surfactant and its interaction with DNA in dilute brine. J. Colloid Interface Sci. 2011, 358, 513–520. [Google Scholar] [CrossRef]
- Karlsson, L.; van Eijk, M.C.; Söderman, O. Compaction of DNA by gemini surfactants: Effects of surfactant architecture. J. Colloid Interface Sci. 2002, 252, 290–296. [Google Scholar] [CrossRef]
- Grueso, E.; Kuliszewska, E.; Roldan, E.; Perez-Tejeda, P.; Rafael Prado-Gotor, R.; Brecker, L. DNA conformational changes induced by cationic gemini surfactants: The key to switching DNA compact structures into elongated forms. RSC Adv. 2015, 5, 29433–29446. [Google Scholar] [CrossRef]
- Grueso, E.; Roldan, E.; Perez-Tejeda, P.; Kuliszewska, E.; Molero, B.; Brecker, L.; Giráldez-Pérez, R.M. Reversible DNA compaction induced by partial intercalation of 16-Ph-16 gemini surfactants: Evidence of triple helix formation. Phys. Chem. Chem. Phys. 2018, 20, 24902–24914. [Google Scholar] [CrossRef] [PubMed]
- Giráldez-Pérez, R.M.; Grueso Molina, E.M.; Lhamyani, S.; Perez Tejeda, M.P.; Gentile, A.M.; Kuliszewska, E.; Roman-Perez, J.; El Bekay, R. miR-21/gemini surfactant-capped gold nanoparticles as potential therapeutic complexes: Synthesis, characterization and in vivo nanotoxicity probes. J. Mol. Liq. 2020, 313, 113577. [Google Scholar] [CrossRef]
- Muddineti, O.S.; Ghosh, B.; Biswas, S. Current trends in using polymer coated gold nanoparticles for cancer therapy. Int. J. Pharm. 2015, 484, 252–267. [Google Scholar] [CrossRef]
- Kennedy, B.J.; Theologides, A. The role of 5-fluorouracil in malignant disease. Ann. Intern Med. 1961, 55, 719–730. [Google Scholar] [CrossRef] [PubMed]
- De la Cueva, A.; Ramírez de Molina, A.; Álvarez-Ayerza, N.; Ramos, M.A.; Cebrián, A.; Gómez Del Pulgar, T.; Lacal, J.C. Combined 5-FU and ChoKa inhibitors as a new alternative therapy of colorectal cancer: Evidence in human tumor-derived cell lines and mouse xenografts. PLoS ONE 2013, 8, e64961. [Google Scholar] [CrossRef] [Green Version]
- Macdonald, J.S. Toxicity of 5-fluorouracil. Oncology 1999, 7 (Suppl. 3), 33–34. [Google Scholar]
- Liszbinski, R.B.; Romagnoli, G.G.; Gorgulho, C.M.; Basso, C.R.; Pedrosa, V.A.; Kaneno, R. Anti-EGFR-coated gold nanoparticles in vitro carry 5-fluorouracil to colorectal cancer cells. Materials 2020, 13, 375. [Google Scholar] [CrossRef] [Green Version]
- Mahdi, W.A.; Hussain, A.; Ramzan, M.; Faruk, A.; Bukhari, S.I.; Dev, A. Pluronic-coated biogenic gold nanoparticles for colon delivery of 5-fluorouracil: In vitro and ex vivo studies. AAPS PharmSciTechnol. 2021, 22, 64. [Google Scholar] [CrossRef]
- Zhao, J.G.; Ren, K.M.; Tang, J. Overcoming 5-Fu resistance in human non-small cell lung cancer cells by the combination of 5-Fu and cisplatin through the inhibition of glucose metabolism. Tumour Biol. 2014, 35, 12305–12315. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, S.; Mahdavi, R.; Alipoor, B.; Panahi, G.; Nasli Esfahani, E.; Razi, F.; Taghikhani, M.; Meshkani, R. Decreased serum microRNA-21 level is associated with obesity in healthy and type 2 diabetic subjects. Arch. Physiol. Biochem. 2018, 124, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold SpringHarbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Felsendeld, G.; Hirschman, S.Z. A neighbor-interaction analysis of the hypochromism and spectra of DNA. J. Mol. Biol. 1965, 13, 407–427. [Google Scholar] [CrossRef]
- Shapiro, D.L.; Nardone, L.L.; Rooney, S.A.; Motoyama, E.K.; Munoz, J.L. Phospholipid biosynthesis and secretion by a cell line (A549) which resembles type II aleveolar epithelial cells. Biochim. Biophys. Acta 1978, 530, 197–207. [Google Scholar] [CrossRef]
- Lieber, M.; Smith, B.; Szakal, A.; Nelson-Rees, W.; Todaro, G.A. Continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int. J. Cancer 1976, 17, 62–70. [Google Scholar] [CrossRef]
- Foster, K.A.; Oster, C.G.; Mayer, M.M.; Avery, M.L.; Audus, K.L. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp. Cell Res. 1998, 243, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Nardone, L.L.; Andrews, S.B. Cell line A549 as a model of the type II pneumocyte: Phospholipid biosynthesis from native and organometallic precursors. Biochim. Biophys. Acta BBA-Lipids Lipid Metab. 1979, 573, 276–295. [Google Scholar] [CrossRef]
- Laschewsky, A.; Lunkenheimer, K.; Rakotoaly, R.H.; Wattebled, L. Spacer effects in dimeric cationic surfactants. Colloid. Polym. Sci. 2005, 283, 469–479. [Google Scholar] [CrossRef]
- Kuliszewska, E.; Brecker, L. Gemini surfactants foam formation ability and foam stability depends on spacer length. J. Surf. Deterg. 2014, 17, 951–957. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, G.; Jin, S.; Xu, L.; Zhao, C.-X. Development of high-drug-loading nanoparticles. ChemPlusChem 2020, 85, 2143–2157. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, D.; Sun, X.; Zeng, S.; Li, L.; Sun, D. Thermodynamic and spectrographic studies on the interactions of ct-DNA with 5-fluorouracil and tegafur. Thermochim. Acta 2009, 493, 30–36. [Google Scholar] [CrossRef]
- Jangir, D.K.; Charak, S.; Mehrotra, R.; Kundu, S. FTIR and circular dichroism spectroscopic study of interaction of 5-fluorouracil with DNA. J. Photochem. Photobiol. B. 2011, 105, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Nucleic Acid Structure and Recognition; Oxford University Press: New York, NY, USA, 2002. [Google Scholar]
- Prado-Gotor, R.; Grueso, E. A kinetic study of the interaction of DNA with gold nanoparticles: Mechanistic aspects of the interaction. Phys. Chem. Chem. Phys. 2011, 13, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Grueso, E.; Giráldez-Pérez, R.M.; Pérez-Tejeda, P.; Roldán, E.; Prado-Gotor, R. What controls the unusual melting profiles of small AuNPs/DNA complexes. Phys.Chem.Chem.Phys. 2019, 21, 11019–11032. [Google Scholar] [CrossRef]
- Xiaofang, Z.; Yazhuo, S.; Honglai, L.; Ying, H. Interaction of DNA with cationic gemini surfactant trimethylene-1,3-bis (dodecyldimethylammonium bromide) and anionic surfactant SDS mixed system. J. Colloid Interface Sci. 2007, 314, 478–483. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of gold nanoparticles from UV−Vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef] [PubMed]
- Zuber, A.; Purdey, M.; Schartner, E.; Forbes, C.; van der Hoekc, B.; Giles, D.; Abell, A.; Monroa, T.; Ebendorff-Heidepriema, H. Detection of gold nanoparticles with different sizes using absorption and fluorescence based method. Sens. Actuators B 2016, 227, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Agyapong, D.A.Y.; Zhao, R.L.; Li, C.X.; Du, W.J.; Zeng, H.J. Novel Mercaptan Derivatives Nanogold Particles with Hyperchromic and Fluorescence Sensitization Effects. In Proceedings of the 2016 International Conference on Biological Sciences and Technology, Guangzhou, China, 8–10 January 2016. [Google Scholar] [CrossRef]
- Freitas, C.; Müller, R.H. Effect of light and temperature on zeta potential and physical stability in solid lipid nanoparticle (SLN (TM)) dispersions. Int. J. Pharm. 1998, 168, 221–229. [Google Scholar] [CrossRef]
- Lee, H.; Mijovíc, J. Bio-nano complexes: DNA/surfactant/single-walled carbon nanotube interactions in electric field. Polymer 2009, 50, 881–890. [Google Scholar] [CrossRef]
- Carnerero, J.M.; Jimenez-Ruiz, A.; Grueso, E.; Prado-Gotor, R. Understanding and improving aggregated gold nanoparticle/dsDNA interactions by molecular spectroscopy and deconvolution methods. Phys.Chem.Chem.Phys 2017, 19, 16113–16123. [Google Scholar] [CrossRef] [PubMed]
- Fuller, M.; Kӧper, I. Polyelectrolyte-coated gold nanoparticles: The effect of salt and polyelectrolyte concentration on colloidal stability. Polymers 2018, 10, 1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.H.; Lee, C.W.; Chiou, A.; Wei, P.-K. Size-dependent endocytosis of gold nanoparticles studied by three-dimensional mapping of plasmonic scattering images. J. Nanobiotechnol. 2010, 8, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nativo, P.; Prior, I.A.; Brust, M. Uptake and intracellular fate of surface-modified gold nanoparticles. ACS Nano 2008, 2, 1639–1644. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.D.; Chan, W.C.W. Elucidating the mechanism of cellar uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Kim, B.Y.S.; Rutka, J.T.; Chan, W.C.W. Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotechnol. 2008, 3, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Alkilany, A.M.; Mahmoud, N.N.; Hashemi, F.; Hajipour, M.J.; Farvadi, F.; Mahmoudi, M. Misinterpretation in nanotoxicology: A personal perspective. Chem. Res. Toxicol. 2016, 29, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Oh, N.; Park, J.H. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int. J. Nanomed. 2014, 9, 51–63. [Google Scholar] [CrossRef] [Green Version]













| Nanosystem | Drug Loading (%) | Drug Release (%) | t1/2 Release (Min) |
|---|---|---|---|
| C1 | 97% | 94% | 1.0 min |
| C2 | 95% | 98 % | 3.98 min |
| C3 | 96% | 99 % | 6.3 min |
| System | Zeta Potential/mV in Water (19×) | Zeta Potential/mV in PBS 0.1× (19×) | Diameter/nm in Water (19×) | Diameter/nm in PBS 0.1× (19×) | Diameter/nm in PBS 0.1× (1×) |
|---|---|---|---|---|---|
| N1 | (30 ± 3) | (18 ± 2) | (13 ± 4) | (23 ± 2) | (23 ± 2) |
| N2 | (35 ± 3) | (28 ± 3) | (10 ± 2) | (21 ± 3) | (23 ± 3) |
| N3 | (40.0 ± 1.0) | (37.0 ± 1.0) | (9 ± 2) | (10 ± 3) | (13 ± 3) |
| C1 | (−30 ± 3) | (−45 ± 8) | (36 ± 6) | (34 ± 7) | d1 = (38 ± 16); 29.7% d2 = (106 ± 13); 70.3% |
| C2 | (−29 ± 6) | (−46 ± 6) | (88 ± 6) | (85 ± 5) | d1 = (41 ± 19); 99.6% d2 = (255 ± 30); 0.4% |
| C3 | (−33 ± 3) | (−56 ± 5) | (16 ± 4) | (14 ± 3) | (26 ± 5) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giráldez-Pérez, R.M.; Grueso, E.; Domínguez, I.; Pastor, N.; Kuliszewska, E.; Prado-Gotor, R.; Requena-Domenech, F. Biocompatible DNA/5-Fluorouracil-Gemini Surfactant-Functionalized Gold Nanoparticles as Promising Vectors in Lung Cancer Therapy. Pharmaceutics 2021, 13, 423. https://doi.org/10.3390/pharmaceutics13030423
Giráldez-Pérez RM, Grueso E, Domínguez I, Pastor N, Kuliszewska E, Prado-Gotor R, Requena-Domenech F. Biocompatible DNA/5-Fluorouracil-Gemini Surfactant-Functionalized Gold Nanoparticles as Promising Vectors in Lung Cancer Therapy. Pharmaceutics. 2021; 13(3):423. https://doi.org/10.3390/pharmaceutics13030423
Chicago/Turabian StyleGiráldez-Pérez, Rosa M., Elia Grueso, Inmaculada Domínguez, Nuria Pastor, Edyta Kuliszewska, Rafael Prado-Gotor, and Francisco Requena-Domenech. 2021. "Biocompatible DNA/5-Fluorouracil-Gemini Surfactant-Functionalized Gold Nanoparticles as Promising Vectors in Lung Cancer Therapy" Pharmaceutics 13, no. 3: 423. https://doi.org/10.3390/pharmaceutics13030423