Anti-Inflammatory Effects of a Polyphenol, Catechin-7,4′-O-Digallate, from Woodfordia uniflora by Regulating NF-κB Signaling Pathway in Mouse Macrophages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Extraction and Isolation
2.2. Cell Culture
2.3. Cell Viability Assay
2.4. Nitric Oxide Assay
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. RNA Extraction and cDNA Synthesis
2.7. Quantitative Polymerase Chain Reaction (qPCR)
2.8. Western Blotting
2.9. Immunofluorescence Technique
2.10. Statistical Analysis
3. Results and Discussions
3.1. Isolation and Identification of Catechin-7,4′-O-Digallate from W. uniflora
3.2. Inhibition of NO Production and iNOS Expression by CDG in LPS-Stimulated ImKCs
3.3. Regulation of Pro-Inflammatory Cytokine Production by CDG in LPS-Stimulated ImKCs
3.4. Inhibition of Phosphorylation and Degradation of IκBα by CDG in LPS-Stimulated ImKCs
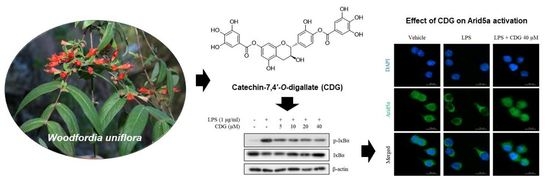
3.5. Inhibition of Arid5a Activation by CDG in LPS-Stimulated ImKCs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fujiwara, N.; Kobayashi, K. Macrophages in inflammation. Curr. Drug Targets 2005, 4, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Koh, T.J.; DiPietro, L.A. Inflammation and wound healing: The role of the macrophage. Expert Rev. Mol. Med. 2011, 13, e23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosmann, M.; Ward, P.A. The inflammatory response in sepsis. Trends Immunol. 2013, 34, 129–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isomäki, P.; Punnonen, J. Pro-and anti-inflammatory cytokines in rheumatoid arthritis. Ann. Med. 1997, 29, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Podolsky, D.K. Inflammatory bowel disease. N. Engl. J. Med. 1991, 325, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Liaskou, E.; Wilson, D.V.; Oo, Y.H. Innate immune cells in liver inflammation. Mediators Inflamm. 2012, 2012, 949157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choy, E.H.; Panayi, G.S. Cytokine pathways and joint inflammation in rheumatoid arthritis. N. Engl. J. Med. 2001, 344, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef] [Green Version]
- Weiming, X.; Liu, L.Z.; Loizidou, M.; Ahmed, M.; Charles, I.G. The role of nitric oxide in cancer. Cell Res. 2002, 12, 311–320. [Google Scholar]
- Aktan, F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004, 75, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Ahmadinejad, F.; Møller, S.G.; Hashemzadeh-Chaleshtori, M.; Bidkhori, G.; Jami, M.S. Molecular Mechanisms behind Free Radical Scavengers Function against Oxidative Stress. Antioxidants 2017, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Nash, K.M.; Schiefer, I.T.; Shah, Z.A. Development of a reactive oxygen species-sensitive nitric oxide synthase inhibitor for the treatment of ischemic stroke. Free Radic. Biol. Med. 2018, 115, 395–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cross, R.K.; Wilson, K.T. Nitric oxide in inflammatory bowel disease. Inflamm. Bowel Dis. 2003, 9, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Dawson, V.L.; Dawson, T.M. Role of nitric oxide in Parkinson’s disease. Pharmacol. Ther. 2006, 109, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.C.; Lee, S.; Kim, S.; Jo, M.S.; Yu, J.S.; Ko, Y.-J.; Cho, Y.-C.; Kim, K.H. Withaninsams A and B: Phenylpropanoid Esters from the Roots of Indian Ginseng (Withania somnifera). Plants 2019, 8, 527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, M.S.; Lee, S.; Yu, J.S.; Baek, S.C.; Cho, Y.-C.; Kim, K.H. Megastigmane Derivatives from the Cladodes of Opuntia humifusa and Their Nitric Oxide Inhibitory Activities in Macrophages. J. Nat. Prod. 2020, 83, 684–692. [Google Scholar] [CrossRef]
- Lee, S.R.; Yi, S.A.; Nam, K.H.; Ryoo, R.; Lee, J.; Kim, K.H. Pantheric Acids A–C from a Poisonous Mushroom, Amanita pantherina, Promote Lipid Accumulation in Adipocytes. J. Nat. Prod. 2019, 82, 3489–3493. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.A.; Park, E.-J.; Lee, D.; Song, J.H.; Lee, H.L.; Kim, K.H.; Kim, Y.; Jung, K.; Kang, K.S.; Yoo, J.-E. Estrogenic Activity of Sanguiin H-6 through Activation of Estrogen Receptor a Coactivator-binding Site. Nat. Prod. Sci. 2019, 25, 28–33. [Google Scholar] [CrossRef]
- Lee, S.; Ryoo, R.; Choi, J.H.; Kim, J.-H.; Kim, S.-H.; Kim, K.H. Trichothecene and tremulane sesquiterpenes from a hallucinogenic mushroom Gymnopilus junonius and their cytotoxicity. Arch. Pharm. Res. 2020, 43, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Kang, H.S.; Yoo, M.J.; Yi, S.A.; Beemelmanns, C.; Lee, J.C.; Kim, K.H. Anti-adipogenic Pregnane Steroid from a Hydractinia-associated Fungus, Cladosporium sphaerospermum SW67. Nat. Prod. Sci. 2020, 26, 230–235. [Google Scholar]
- Yu, J.S.; Park, M.; Pang, C.; Rashan, L.; Jung, W.H.; Kim, K.H. Antifungal Phenols from Woodfordia uniflora Collected in Oman. J. Nat. Prod. 2020, 83, 2261–2268. [Google Scholar] [CrossRef] [PubMed]
- El-Toumy, S.A.A.; Mahdy, K.A. Polyphenols from Acacia nilotica leaves and evaluation of antihyperglycaemic effect of aqueous extract. Bull. Fac. Pharm Cairo Univ. 2004, 42, 317–325. [Google Scholar]
- Eberhardt, M.V.; Lee, C.Y.; Liu, R.H. Antioxidant activity of fresh apples. Nature 2000, 405, 903–904. [Google Scholar] [CrossRef]
- Recio, M.C.; Andujar, I.; Rios, J.L. Anti-inflammatory agents from plants: Progress and potential. Curr. Med. Chem. 2012, 19, 2088–2103. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo, C.; Russo, M.; Bilotto, S.; Tedesco, I.; Laratta, B.; Russo, G.L. Dietary polyphenols in cancer prevention: The example of the flavonoid quercetin in leukemia. Ann. N. Y. Acad. Sci. 2012, 1259, 95–103. [Google Scholar] [CrossRef]
- Andriantsitohaina, R.; Auger, C.; Chataigneau, T.; Étienne-Selloum, N.; Li, H.; Martínez, M.C.; Schini-Kerth, V.B.; Laher, I. Molecular mechanisms of the cardiovascular protective effects of polyphenols. Br. J. Nutr. 2012, 108, 1532–1549. [Google Scholar] [CrossRef] [Green Version]
- Vauzour, D.; Rodriguez-Mateos, A.; Corona, G.; Oruna-Concha, M.J.; Spencer, J.P. Polyphenols and human health: Prevention of disease and mechanisms of action. Nutrients 2010, 2, 1106–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tipoe, G.L.; Leung, T.-M.; Hung, M.-W.; Fung, M.-L. Green tea polyphenols as an anti-oxidant and anti-inflammatory agent for cardiovascular protection. Cardiovasc. Hematol. Disord. Drug Targets 2007, 7, 135–144. [Google Scholar] [CrossRef]
- Wojdasiewicz, P.; Poniatowski, L.A.; Szukiewicz, D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediat. Inflamm. 2014, 2014, 561459. [Google Scholar] [CrossRef] [Green Version]
- Ivashkiv, L.B. Inflammatory signaling in macrophages: Transitions from acute to tolerant and alternative activation states. Eur. J. Immunol. 2011, 41, 2477–2481. [Google Scholar] [CrossRef] [Green Version]
- Li, M.-Y.; Sun, L.; Niu, X.-T.; Chen, X.-M.; Tian, J.-X.; Kong, Y.-D.; Wang, G.-Q. Astaxanthin protects lipopolysaccharide-induced inflammatory response in Channa argus through inhibiting NF-κB and MAPKs signaling pathways. Fish Shellfish Immunol. 2019, 86, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Tak, P.P.; Firestein, G.S. NF-κB: A key role in inflammatory diseases. J. Clin. Invest. 2001, 107, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Kashiwabara, Y.; Nathan, C. Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. J. Biol. Chem. 1994, 269, 4705–4708. [Google Scholar] [CrossRef]
- Yeom, M.; Kim, J.-H.; Min, J.-H.; Hwang, M.K.; Jung, H.-S.; Sohn, Y. Xanthii fructus inhibits inflammatory responses in LPS-stimulated RAW 264.7 macrophages through suppressing NF-κB and JNK/p38 MAPK. J. Ethnopharmacol. 2015, 176, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.G.; Jin, H.; Yu, P.J.; Tian, Y.X.; Zhang, J.J.; Wu, S.G. Mollugin inhibits the inflammatory response in lipopolysaccharide-stimulated RAW264.7 macrophages by blocking the Janus kinase-signal transducers and activators of transcription signaling pathway. Biol. Pharm. Bull. 2013, 36, 399–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, J.; Ciesielski, C.J.; Hunt, A.E.; Horwood, N.J.; Beech, J.T.; Hayes, L.A.; Denys, A.; Feldmann, M.; Brennan, F.M.; Foxwell, B.M. A novel mechanism for TNF-α regulation by p38 MAPK: Involvement of NF-κB with implications for therapy in rheumatoid arthritis. J. Immunol. 2004, 173, 6928–6937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Johnson, V.J.; Sharma, R.P. Mercury inhibits nitric oxide production but activates proinflammatory cytokine expression in murine macrophage: Differential modulation of NF-κB and p38 MAPK signaling pathways. Nitric Oxide 2002, 7, 67–74. [Google Scholar] [CrossRef]
- Hasan, A.; Cotobal, C.; Duncan, C.D.; Mata, J. Systematic analysis of the role of RNA-binding proteins in the regulation of RNA stability. PLoS Genet. 2014, 10, e1004684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Liu, M.; D’Silva, N.J.; Kirkwood, K.L. Tristetraprolin regulates interleukin-6 expression through p38 MAPK-dependent affinity changes with mRNA 3′ untranslated region. J. Interf. Cytokine Res. 2011, 31, 629–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.-X.; Wang, C.; Huang, S.-F.; Chen, Q.; Hu, Y.-F.; Zhou, L.; Gu, Y. Regnase-1 in microglia negatively regulates high mobility group box 1-mediated inflammation and neuronal injury. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Higa, M.; Oka, M.; Fujihara, Y.; Masuda, K.; Yoneda, Y.; Kishimoto, T. Regulation of inflammatory responses by dynamic subcellular localization of RNA-binding protein Arid5a. Proc. Natl. Acad. Sci. USA 2018, 115, E1214–E1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyati, K.K.; Agarwal, R.G.; Sharma, P.; Kishimoto, T. Arid5a regulation and the roles of Arid5a in the inflammatory response and disease. Front. Immunol. 2019, 10, 2790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyati, K.K.; Masuda, K.; Zaman, M.M.-U.; Dubey, P.K.; Millrine, D.; Chalise, J.P.; Higa, M.; Li, S.; Standley, D.M.; Saito, K. TLR4-induced NF-κB and MAPK signaling regulate the IL-6 mRNA stabilizing protein Arid5a. Nucleic Acids Res. 2017, 45, 2687–2703. [Google Scholar] [CrossRef]
- Masuda, K.; Ripley, B.; Nishimura, R.; Mino, T.; Takeuchi, O.; Shioi, G.; Kiyonari, H.; Kishimoto, T. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 9409–9414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyati, K.K.; Zaman, M.M.-U.; Sharma, P.; Kishimoto, T. Arid5a, an RNA-binding protein in immune regulation: RNA stability, inflammation, and autoimmunity. Trends Immunol. 2020, 41, 255–268. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.J.; Seo, J.B.; Yu, J.S.; Lee, S.; Lim, J.S.; Choi, J.U.; Lee, C.-M.; Rashan, L.; Kim, K.H.; Cho, Y.-C. Anti-Inflammatory Effects of a Polyphenol, Catechin-7,4′-O-Digallate, from Woodfordia uniflora by Regulating NF-κB Signaling Pathway in Mouse Macrophages. Pharmaceutics 2021, 13, 408. https://doi.org/10.3390/pharmaceutics13030408
Kim EJ, Seo JB, Yu JS, Lee S, Lim JS, Choi JU, Lee C-M, Rashan L, Kim KH, Cho Y-C. Anti-Inflammatory Effects of a Polyphenol, Catechin-7,4′-O-Digallate, from Woodfordia uniflora by Regulating NF-κB Signaling Pathway in Mouse Macrophages. Pharmaceutics. 2021; 13(3):408. https://doi.org/10.3390/pharmaceutics13030408
Chicago/Turabian StyleKim, Eui Jin, Ji Bin Seo, Jae Sik Yu, Seoyoung Lee, Jae Sung Lim, Jeong Uk Choi, Chang-Min Lee, Luay Rashan, Ki Hyun Kim, and Young-Chang Cho. 2021. "Anti-Inflammatory Effects of a Polyphenol, Catechin-7,4′-O-Digallate, from Woodfordia uniflora by Regulating NF-κB Signaling Pathway in Mouse Macrophages" Pharmaceutics 13, no. 3: 408. https://doi.org/10.3390/pharmaceutics13030408