Novel Green Biosynthesis of 5-Fluorouracil Chromium Nanoparticles Using Harpullia pendula Extract for Treatment of Colorectal Cancer
Abstract
:1. Introduction
2. Material and Methods
2.1. Materials
2.2. Methods
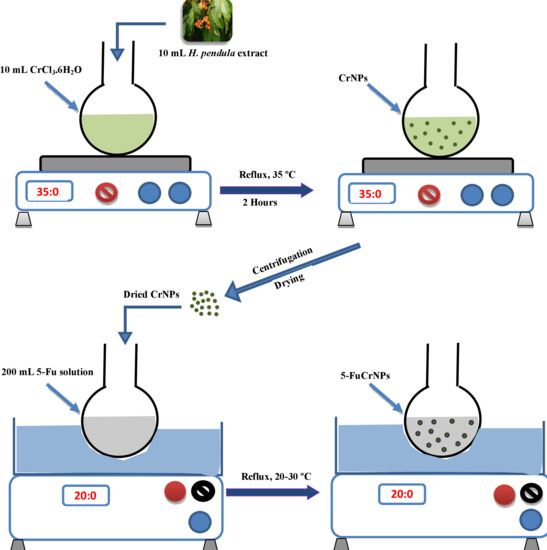
2.2.1. Plant Material and Extract Preparation
2.2.2. Biosynthesis of Chromium Nanoparticles (CrNPs)
2.2.3. Preparation of 5-Fu Loaded Chromium Nanoparticles (5-FuCrNPs)
2.2.4. Characterization of the Prepared NPs.
X-ray Powder Diffraction (XRD)
Fourier Transform Infrared Spectroscopy (FTIR)
Scanning Electron Microscopy (SEM)
Transmission Electron Microscopy (TEM)
Determination of Drug Loading Efficiency Percentage (LE%)
2.2.5. In Vitro Drug Release Study
Experimental Design (BBD)
2.2.6. Effect on Cell Proliferation
3. Results and Discussion
3.1. XRD Diffraction of the Biosynthesized CrNPs
3.2. SEM and TEM Analyses of Biosynthesized CrNPs and 5-FuCrNPs
3.3. Functional Groups Identification Utilizing FTIR Spectroscopy
3.4. Drug Loading Efficiency on CrNPs (LE%)
3.5. In Vitro Release Study of 5-FuCrNPs
3.6. In Vitro Cytotoxicity Assay
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gast. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, M.P.; Choong, K.C.; Thornblade, L.W.; Fakih, M.G.; Fong, Y.; Kaiser, A.M. Management considerations for the surgical treatment of colorectal Cancer during the global Covid-19 pandemic. Ann. Surg. 2020, 272, e98–e105. [Google Scholar] [CrossRef] [PubMed]
- Schuhmacher, C.; Gretschel, S.; Lordick, F.; Reichardt, P.; Hohenberger, W.; Eisenberger, C.F.; Haag, C.; Mauer, M.E.; Hasan, B.; Welch, J. Neoadjuvant chemotherapy compared with surgery alone for locally advanced cancer of the stomach and cardia: European Organisation for Research and Treatment of Cancer randomized trial 40954. J. Clin. Oncol. 2010, 28, 5210–5218. [Google Scholar] [CrossRef]
- Baskar, R.; Lee, K.A.; Yeo, R.; Yeoh, K.-W. Cancer and radiation therapy: Current advances and future directions. Int. J. Med Sci. 2012, 9, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Rebucci, M.; Michiels, C. Molecular aspects of cancer cell resistance to chemotherapy. Biochem. Pharmacol. 2013, 85, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Vodenkova, S.; Buchler, T.; Cervena, K.; Veskrnova, V.; Vodicka, P.; Vymetalkova, V. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharm. Therap. 2020, 206, 107447. [Google Scholar] [CrossRef]
- de Gramont, A.d.; Figer, A.; Seymour, M.; Homerin, M.; Hmissi, A.; Cassidy, J.; Boni, C.; Cortes-Funes, H.; Cervantes, A.; Freyer, G. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J. Clin. Oncol. 2000, 18, 2938–2947. [Google Scholar] [CrossRef]
- Nair, L.; Jagadeeshan, S.; Nair, S.A.; Kumar, G.V. Biological evaluation of 5-fluorouracil nanoparticles for cancer chemotherapy and its dependence on the carrier, PLGA. Int. J. Nanomed. 2011, 6, 1685–1697. [Google Scholar] [CrossRef] [Green Version]
- Braun, M.S.; Seymour, M.T. Balancing the efficacy and toxicity of chemotherapy in colorectal cancer. Ther. Adv. Med Oncol. 2010, 3, 43–52. [Google Scholar] [CrossRef] [Green Version]
- He, Y.-C.; Chen, J.-W.; Cao, J.; Pan, D.-Y.; Qiao, J.-G. Toxicities and therapeutic effect of 5-fluorouracil controlled release implant on tumor-bearing rats. World J. Gastroenterol. 2003, 9, 1795–1798. [Google Scholar] [CrossRef]
- Wilkinson, J. Nanotechnology applications in medicine. Med. Dev. Technol. 2003, 14, 29. [Google Scholar]
- Jameel, A.T.; Yaser, A.Z. Advances in Nanotechnology and Its Applications; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Sun, Q.; Zhou, Z.; Qiu, N.; Shen, Y. Rational design of cancer nanomedicine: Nanoproperty integration and synchronization. Adv. Mater. 2017, 29, 1606628. [Google Scholar] [CrossRef]
- Qiao, Y.; He, J.; Chen, W.; Yu, Y.; Li, W.; Du, Z.; Xie, T.; Ye, Y.; Hua, S.Y.; Zhong, D. Light-Activatable Synergistic Therapy of Drug-Resistant Bacteria-Infected Cutaneous Chronic Wounds and Nonhealing Keratitis by Cupriferous Hollow Nanoshells. ACS Nano. 2020, 14, 3299–3315. [Google Scholar] [CrossRef]
- Saddik, M.S.; Mohamed, E.E.; Elmahdy, M.M. Preparation and Characterization of Niosomal Carrier System of Hydrophilic Drug (Methylene Blue) for Photodynamic Therapy. Lat. Americ J. Pharm. 2020, 39, 561–569. [Google Scholar]
- Angelova, A.; Angelov, B. Dual and multi-drug delivery nanoparticles towards neuronal survival and synaptic repair. Neural Regen. Res. 2017, 12, 886–889. [Google Scholar] [CrossRef]
- Anderson, R.A. Chromium as an essential nutrient for humans. Regul. Toxicol. Pharmacol. 1997, 26, S35–S41. [Google Scholar] [CrossRef] [Green Version]
- Anderson, R.A. Effects of chromium on body composition and weight loss. Nutr. Rev. 1998, 56, 266–270. [Google Scholar] [CrossRef]
- Sukumar, C.; Gowthami, G.; Nitya, R.; Janaki, V.; Kamala-Kannan, S.; Shanthi, K. Significance of co-immobilized activated carbon and Bacillus subtilis on removal of Cr (VI) from aqueous solutions. Environ. Earth Sci. 2013, 72, 839–847. [Google Scholar] [CrossRef]
- Medina, C.; Santos-Martinez, M.; Radomski, A.; Corrigan, O.; Radomski, M. Nanoparticles: Pharmacological and toxicological significance. Br. J. Pharm. 2007, 150, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Chen, Z.G.; Shin, D.M. Advances of cancer therapy by nanotechnology. Cancer Res. Treat. 2009, 41, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Refaat, H.; Naguib, Y.W.; Elsayed, M.; Sarhan, H.A.; Alaaeldin, E. Modified spraying technique and response surface methodology for the preparation and optimization of propolis liposomes of enhanced anti-proliferative activity against human melanoma cell line A375. Pharmaceutics 2019, 11, 558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auda, S.H.; Abd El-Rasoul, S.; Ahmed, M.M.; Osman, S.K.; El-Badry, M. In-vitro release and in-vivo performance of tolmetin from different topical gel formulations. J. Pharm. Investig. 2015, 45, 311–317. [Google Scholar] [CrossRef]
- Satgurunathan, T.; Bhavan, P.S.; Joy, R.D.S. Green Synthesis of Chromium Nanoparticles and Their Effects on the Growth of the Prawn Macrobrachium rosenbergii Post-larvae. Biol. Trace Element Res. 2018, 187, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Saddik, M.S.; Alsharif, F.M.; El-Mokhtar, M.A.; Al-Hakkani, M.F.; El-Mahdy, M.M.; Farghaly, H.S.; Abou-Taleb, H.A. Biosynthesis, Characterization, and Wound-Healing Activity of Phenytoin-Loaded Copper Nanoparticles. AAPS PharmSciTech 2020, 21, 1–12. [Google Scholar] [CrossRef]
- Kumar, R.; Ghoshal, G.; Jain, A.; Goyal, M. Rapid green synthesis of silver nanoparticles (AgNPs) using (Prunus persica) plants extract: Exploring its antimicrobial and catalytic activities. J. Nanomed. Nanotechnol. 2017, 8, 1–8. [Google Scholar]
- Abboud, Y.; Saffaj, T.; Chagraoui, A.; El Bouari, A.; Brouzi, K.; Tanane, O.; Ihssane, B. Biosynthesis, characterization and antimicrobial activity of copper oxide nanoparticles (CONPs) produced using brown alga extract (Bifurcaria bifurcata). Appl. Nanosci. 2013, 4, 571–576. [Google Scholar] [CrossRef] [Green Version]
- Abd El Rasoul, S.; Saleh, K. Emulsion solvent evaporation method for preparing Eudragit RS100 microparticles loaded ketorolac tromethamine. Asian J. Pharm. Health Sci. 2013, 3, 627–639. [Google Scholar]
- Dizes, C.; Gerald, F.; Lavaud, C.; Ellas, R.; Faure, R.; Massiot, G.; Balansard, G. Harpuloside a triterpenoid saponin from Harpullia ramiflora. Phytochemistry 1998, 48, 1229–1232. [Google Scholar] [CrossRef]
- Poovapatthanachart, R. Phytochemical Study of Harpullia Arborea Leaves. Master’s Thesis, Chulalongkorn University, Bangkok, Thailand, 2003. [Google Scholar]
- Abdelkader, M.S.A.; Rateb, M.E.; Mohamed, G.A.; Jaspars, M. Harpulliasides A and B: Two new benzeneacetic acid derivatives from Harpullia pendula. Phytochem. Lett. 2016, 15, 131–135. [Google Scholar] [CrossRef]
- Ghaly, N.S.; Nabil, M.; Grace, M.H.; Melek, F.R. Pendulaosides A and B. Two acylated triterpenoid saponins from Harpullia pendula seed extract. Phytochem. Lett. 2017, 21, 278–282. [Google Scholar] [CrossRef]
- El Souda, S.S.; Mohammed, R.S.; Ibrahim, F.M.; Matloub, A.A. Harpullia pendula Planch leaves: Phenolics, in vitro antioxidant and α-amylase inhibitory activity. Egypt. Pharm. J. 2017, 16, 103. [Google Scholar] [CrossRef]
- El-Rasoul, A.; Ahmed, M.M. Chitosan polymer as a coat of calcium alginate microcapsules loaded by non-steroidal antiinflammatory drug. Bull. Pharm. Sci. 2010, 33, 179–186. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, M.M.; Abd El-Rasoul, S.; Auda, S.H.; Ibrahim, M.A. Emulsification/internal gelation as a method for preparation of diclofenac sodium–sodium alginate microparticles. Saud Pharm. J. 2013, 21, 61–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsayed, M.M.; Abd El Rasoul, S.; Hussein, A.K. Response Surface Methodology as a Useful Tool for Development and Optimization of Sustained Release Ketorolac Tromethamine Niosomal Organogels. J. Pharm. Innov. 2020, 15, 664–677. [Google Scholar] [CrossRef]
- Marques, S.S.; Ramos, I.I.; Fernandes, S.R.; Barreiros, L.; Lima, S.A.; Reis, S.; Domingues, M.R.M.; Segundo, M.A. Insights on Ultrafiltration-Based Separation for the Purification and Quantification of Methotrexate in Nanocarriers. Molecules 2020, 25, 1879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Luz-Asunción, M.; Sánchez-Mendieta, V.; Martínez-Hernández, A.L.; Castaño, V.; Velasco-Santos, C. Adsorption of phenol from aqueous solutions by carbon nanomaterials of one and two dimensions: Kinetic and equilibrium studies. J. Nanomat. 2015, 2015, 405036. [Google Scholar] [CrossRef] [Green Version]
- Elsayed, M.M.; Mostafa, M.E.; Alaaeldin, E.; Sarhan, H.A.; Shaykoon, M.S.; Allam, S.; Ahmed, A.R.; Elsadek, B.E. Design and characterisation of novel Sorafenib-loaded carbon nanotubes with distinct tumour-suppressive activity in hepatocellular carcinoma. Int. J. Nanomed. 2019, 14, 8445. [Google Scholar] [CrossRef] [Green Version]
- Yetilmezsoy, K.; Demirel, S.; Vanderbei, R.J. Response surface modeling of Pb (II) removal from aqueous solution by Pistacia vera L.: Box–Behnken experimental design. J. Hazard. Mater. 2009, 171, 551–562. [Google Scholar] [CrossRef]
- Ahmed, M.M. Effect of different formulation factors on release characteristics of gastro-floating microspheres of ethyl cellulose/carbopol 934P encapsulating sorafenib. Int. J. Pharm. Pharm. Sci. 2019, 11, 64–70. [Google Scholar]
- El-Shenawy, A.A.; Ahmed, M.M.; Mansour, H.F.; El Rasoul, S.A. Torsemide Fast Dissolving Tablets: Development, Optimization Using Box–Bhenken Design and Response Surface Methodology, In Vitro Characterization, and Pharmacokinetic Assessment. AAPS PharmSciTech 2017, 18, 2168–2179. [Google Scholar] [CrossRef]
- Elsayed, M. Design and optimization of tolmetin sodium microspheres prepared by emulsification-internal gelation using response surface methodology. Al Azh. J. Pharm. Sci. 2012, 45, 383–398. [Google Scholar]
- Motwani, S.K.; Chopra, S.; Talegaonkar, S.; Kohli, K.; Ahmad, F.J.; Khar, R.K. Chitosan–sodium alginate nanoparticles as submicroscopic reservoirs for ocular delivery: Formulation, optimisation and in vitro characterisation. Eur. J. Pharm. Biopharm. 2007, 68, 513–525. [Google Scholar] [CrossRef]
- Sofidiya, M.; Jimoh, F.; Aliero, A.; Afolayan, A.; Odukoya, O.; Familoni, O. Antioxidant and antibacterial properties of Lecaniodiscus cupanioides. Planta Med. 2007, 73, 212. [Google Scholar] [CrossRef]
- Simpson, B.; Claudie, D.; Smith, N.; Wang, J.; McKinnon, R.; Semple, S. Evaluation of the anti-inflammatory properties of Dodonaea polyandra, a Kaanju traditional medicine. J. Ethnopharmacol. 2010, 132, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Díaz, M.; Rossini, C. Bioactive natural products from Sapindaceae deterrent and toxic metabolites against insects. In Insecticides–Pest Engineering; Perveen, F., Ed.; InTech: Rijeka, Croatia, 2012; pp. 287–308. [Google Scholar]
- Muthukumran, P.; Begumand, V.H.; Kalaiarasan, P. Anti-diabetic activity of Dodonaea viscosa (L) leaf extracts. Int. J. Pharm. Tech. Res. 2011, 3, 136–139. [Google Scholar]
- Abdel-Rahman, L.H.; Abu-Dief, A.M.; El-Khatib, R.M.; Abdel-Fatah, S.M. Some new nano-sized Fe (II), Cd (II) and Zn (II) Schiff base complexes as precursor for metal oxides: Sonochemical synthesis, characterization, DNA interaction, in vitro antimicrobial and anticancer activities. Bioorganic Chem. 2016, 69, 140–152. [Google Scholar] [CrossRef]
- Abu-Dief, A.M.; Mohamed, W. α-Bi2O3 nanorods: Synthesis, characterization and UV-photocatalytic activity. Mat. Res. Exp. 2017, 4, 035039. [Google Scholar] [CrossRef]
- Abdel Rahman, L.H.; Abu-Dief, A.M.; El-Khatib, R.M.; Abdel-Fatah, S.M.; Adam, A.; Ibrahim, E. Sonochemical synthesis, structural inspection and semiconductor behavior of three new nano sized Cu (II), Co (II) and Ni (II) chelates based on tri-dentate NOO imine ligand as precursors for metal oxides. Appl. Org. Chem. 2018, 32, e4174. [Google Scholar] [CrossRef]
- Mohamed, W.; Abu-Dief, A.M. Synthesis, characterization and photocatalysis enhancement of Eu2O3-ZnO mixed oxide nanoparticles. J. Phys. Chem. Solids 2018, 116, 375–385. [Google Scholar] [CrossRef]
- Hassanien, A.; Akl, A.A.; Sáaedi, A. Synthesis, crystallography, microstructure, crystal defects, and morphology of BixZn1−xO nanoparticles prepared by sol–gel technique. Cryst. Eng. Comm. 2018, 20, 1716–1730. [Google Scholar] [CrossRef]
- Akl, A.A.; Mahmoud, S.A.; Al-Shomar, S.; Hassanien, A. Improving microstructural properties and minimizing crystal imperfections of nanocrystalline Cu2O thin films of different solution molarities for solar cell applications. Mater. Sci. Semicond. Process. 2018, 74, 183–192. [Google Scholar] [CrossRef]
- Yogamalar, R.; Srinivasan, R.; Vinu, A.; Ariga, K.; Bose, A.C. X-ray peak broadening analysis in ZnO nanoparticles. Solid State Commun. 2009, 149, 1919–1923. [Google Scholar] [CrossRef]
- Jayaseelan, C.; Rahuman, A.A.; Kirthi, A.V.; Marimuthu, S.; Santhoshkumar, T.; Bagavan, A.; Gaurav, K.; Karthik, L.; Rao, K.B. Novel microbial route to synthesize ZnO nanoparticles using Aeromonas hydrophila and their activity against pathogenic bacteria and fungi. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 90, 78–84. [Google Scholar] [CrossRef]
- Sanghi, R.; Verma, P. A facile green extracellular biosynthesis of CdS nanoparticles by immobilized fungus. Chem. Eng. J. 2009, 155, 886–891. [Google Scholar] [CrossRef]
- Masood, F.; Malik, A. Biosorption of metal ions from aqueous solution and tannery effluent by Bacillus sp. FM1. J. Environ. Sci Health Part A 2011, 46, 1667–1674. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.N.; Lakkadwala, S.; Majrad, M.S.; Injeti, E.R.; Gollmer, S.M.; Shah, Z.A.; Boddu, S.H.S.; Nesamony, J. Characterization and evaluation of 5-fluorouracil-loaded solid lipid nanoparticles prepared via a temperature-modulated solidification technique. AAPS PharmSciTech 2014, 15, 1498–1508. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Jin, L.; Chen, X. The effect and prediction of temperature on adsorption capability of coal/CH4. Procedia Eng. 2011, 26, 126–131. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.L.; Dalal, N.S. Chromium (V) and hydroxyl radical formation during the glutathione reductase-catalyzed reduction of chromium (VI). Biochem. Biophys. Res. Commun. 1989, 163, 627–634. [Google Scholar] [CrossRef]
- Collins, A.R. The comet assay for DNA damage and repair: Principles, applications, and limitations. Mol. Biotechnol. 2004, 26, 249–261. [Google Scholar] [CrossRef]
- Alarifi, S.; Ali, D.; Alkahtani, S. Mechanistic investigation of toxicity of chromium oxide nanoparticles in murine fibrosarcoma cells. Int. J. Nanomed. 2016, 11, 1253–1259. [Google Scholar] [CrossRef] [Green Version]
- Xia, T.; Kovochich, M.; Liong, M.; Zink, J.I.; Nel, A.E. Cationic polystyrene nanosphere toxicity depends on cell-specific endocytic and mitochondrial injury pathways. ACS Nano 2008, 2, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Saxena, R.; Singh, S. Chromium induced lipid peroxidation in the plants of Pistia stratiotes L.: Role of antioxidants and antioxidant enzymes. Chemosphere 2005, 58, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Rungby, J.; Ernst, E. Experimentally induced lipid peroxidation after exposure to chromium, mercury or silver: Interactions with carbon tetrachloride. Pharmacol. Toxicol. 1992, 70, 205–207. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Chen, P.; Lu, J.; Zhu, Y.; Xu, Y.; Liu, Y.; Liu, J. Protective Effect of Purple Tomato Anthocyanidin on Chromium(VI)-Induced Autophagy in LMH Cells by Inhibiting Endoplasmic Reticulum Stress. Biol. Trace Element Res. 2019, 194, 570–580. [Google Scholar] [CrossRef]










| Formula No. | The Variable Level in a Coded Form | LE% | Cumulative Percentage Released | |||
|---|---|---|---|---|---|---|
| X1 5-Fu Conc. | X2 CrNP Weight | X3 Temperature | Y1 LE % | Y2 Rel 1 h | Y3 Rel 3 h | |
| N1 | −1 | −1 | 0 | 55.87 ± 2.03 | 14.88 ± 1.88 | 62.15 ± 1.96 |
| N2 | 0 | −1 | −1 | 75.64 ± 3.98 | 18.26 ± 1.93 | 81.77 ± 1.11 |
| N3 | 0 | −1 | 1 | 67.31 ± 2.23 | 24.12 ± 1.77 | 90.77 ± 1.72 |
| N4 | 1 | −1 | 0 | 74.32 ± 2.08 | 21.90 ± 1.65 | 79.42 ± 1.33 |
| N5 | −1 | 0 | −1 | 80.25 ± 2.21 | 11.89 ± 1.98 | 76.09 ± 1.94 |
| N6 | −1 | 0 | 1 | 67.76 ± 3.05 | 9.83 ± 1.34 | 68.13 ± 1.56 |
| N7 | 0 | 0 | 0 | 74.13 ± 2.06 | 18.14 ± 1.65 | 87.11 ± 1.65 |
| N8 | 0 | 0 | 0 | 83.23 ± 4.12 | 14.24 ± 1.74 | 84.51 ± 1.44 |
| N9 | 0 | 0 | 0 | 80.99 ± 3.11 | 14.02 ± 1.83 | 82.93 ± 1.76 |
| N10 | 1 | 0 | −1 | 92.11 ± 3.23 | 18.33 ± 1.43 | 91.23 ± 1.34 |
| N11 | 1 | 0 | 1 | 81.91 ± 3.87 | 16.36 ± 1.87 | 92.73 ± 1.33 |
| N12 | −1 | 1 | 0 | 74.33 ± 2.99 | 9.88 ± 1.96 | 71.45 ± 1.93 |
| N13 | 0 | 1 | −1 | 85.89 ± 2.01 | 16.02 ± 1.54 | 80.69 ± 1.25 |
| N14 | 0 | 1 | 1 | 76.81 ± 3.43 | 18.61 ± 1.66 | 69.93 ± 1.54 |
| N15 | 1 | 1 | 0 | 87.54 ± 2.02 | 15.92 ± 1.99 | 74.18 ± 1.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saddik, M.S.; Elsayed, M.M.A.; Abdelkader, M.S.A.; El-Mokhtar, M.A.; Abdel-Aleem, J.A.; Abu-Dief, A.M.; Al-Hakkani, M.F.; Farghaly, H.S.; Abou-Taleb, H.A. Novel Green Biosynthesis of 5-Fluorouracil Chromium Nanoparticles Using Harpullia pendula Extract for Treatment of Colorectal Cancer. Pharmaceutics 2021, 13, 226. https://doi.org/10.3390/pharmaceutics13020226
Saddik MS, Elsayed MMA, Abdelkader MSA, El-Mokhtar MA, Abdel-Aleem JA, Abu-Dief AM, Al-Hakkani MF, Farghaly HS, Abou-Taleb HA. Novel Green Biosynthesis of 5-Fluorouracil Chromium Nanoparticles Using Harpullia pendula Extract for Treatment of Colorectal Cancer. Pharmaceutics. 2021; 13(2):226. https://doi.org/10.3390/pharmaceutics13020226
Chicago/Turabian StyleSaddik, Mohammed S., Mahmoud M. A. Elsayed, Mohamed Salaheldin A. Abdelkader, Mohamed A. El-Mokhtar, Jelan A. Abdel-Aleem, Ahmed M. Abu-Dief, Mostafa F. Al-Hakkani, Hatem S. Farghaly, and Heba A. Abou-Taleb. 2021. "Novel Green Biosynthesis of 5-Fluorouracil Chromium Nanoparticles Using Harpullia pendula Extract for Treatment of Colorectal Cancer" Pharmaceutics 13, no. 2: 226. https://doi.org/10.3390/pharmaceutics13020226