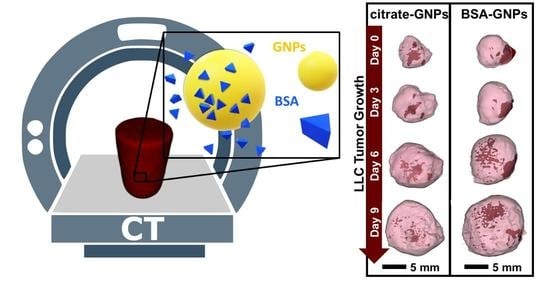
Effects of Surface Protein Adsorption on the Distribution and Retention of Intratumorally Administered Gold Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Gold Nanoparticles
2.2. Surface Passivation of Gold Nanoparticles
2.3. Characterization and Physicochemical Properties of Gold Nanoparticles
2.4. BSA Adsorption on Gold Nanoparticles
2.4.1. Preparation of BSA-GNP Conjugates at Different pH Values
2.4.2. Adsorption Model
2.4.3. Characterization in Various Media
2.5. Cellular Uptake of GNPs and Cytotoxicity In Vitro
2.5.1. Maintenance and LLC Subculture
2.5.2. Trypan Blue Assay
2.5.3. Quantification of Intracellular Gold Content Using ICP-OES
2.5.4. MTT Assay for Cytotoxicity
2.5.5. Scanning Transmission Electron Microscopy (STEM) to Confirm GNP Uptake
2.6. In Vivo Biodistribution and Retention of GNPs
2.6.1. C57BL/6 Mice and LLC Model
2.6.2. Determination of Au in Organs and Blood
2.7. Statistical Analysis
3. Results
3.1. GNP Characterization and Physicochemical (Charge, Size, Functionalization, X-ray Attenuation) Properties
3.2. BSA Adsorption Models on GNPs
3.3. In Vitro Uptake of GNPs
3.4. In Vivo Biodistribution and Retention of GNPs
3.4.1. Manual Intratumoral Injection of Low Dose GNPs
3.4.2. Automatic Syringe Pump Intratumoral Injection of High Dose GNPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | analysis of variance |
| BSA | Bovine Serum Albumin |
| CT | Computed Tomography |
| DLS | Dynamic light scattering |
| EPR | enhanced permeability and retention effect |
| FBS | fetal bovine serum |
| GNP | gold nanoparticle |
| HU | Hounsfield unit |
| IACUC | Institutional Animal Care and Use Committee |
| IAW | Inveon Acquisition Workplace |
| ICP-OES | Inductively Coupled Plasma Optical Emission Spectrometry |
| IT | intra-tumoral |
| LLC | Lewis Lung Carcinoma |
| LLC-Luc | luciferase expressing Lewis Lung Carcinoma |
| MM | Multi-Modality |
| MTT | (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) |
| NSCLC | Non-Small Cell Lung Cancer |
| OsO4 | osmium tetroxide |
| PBS | phosphate buffered saline |
| PDI | polydispersity index |
| pI | Isoelectric Point |
| ROI | region of interest |
| s.e.m. | standard error of the mean |
| SERS | Surface-enhanced Raman scattering |
| SPR | surface plasmon resonance |
| STEM | Scanning transmission electron microscopy |
References
- Sumer, B.; Gao, J. Theranostic Nanomedicine for Cancer. Nanomedicine 2008, 3, 137–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chua, C.Y.X.; Ho, J.; Demaria, S.; Ferrari, M.; Grattoni, A. Emerging Technologies for Local Cancer Treatment. Adv. Therap. 2020, 3, 2000027. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Dilnawaz, F.; Sahoo, S.K. Challenges of Moving Theranostic Nanomedicine into the Clinic. Nanomedicine 2020, 15, 111–114. [Google Scholar] [CrossRef]
- Auría-Soro, C.; Nesma, T.; Juanes-Velasco, P.; Landeira-Viñuela, A.; Fidalgo-Gomez, H.; Acebes-Fernandez, V.; Gongora, R.; Almendral Parra, M.J.; Manzano-Roman, R.; Fuentes, M. Interactions of Nanoparticles and Biosystems: Microenvironment of Nanoparticles and Biomolecules in Nanomedicine. Nanomaterials 2019, 9, 1365. [Google Scholar] [CrossRef] [Green Version]
- Vasti, C.; Bedoya, D.A.; Rojas, R.; Giacomelli, C.E. Effect of the Protein Corona on the Colloidal Stability and Reactivity of LDH-Based Nanocarriers. J. Mater. Chem. B 2016, 4, 2008–2016. [Google Scholar] [CrossRef]
- Gebauer, J.S.; Malissek, M.; Simon, S.; Knauer, S.K.; Maskos, M.; Stauber, R.H.; Peukert, W.; Treuel, L. Impact of the Nanoparticle–Protein Corona on Colloidal Stability and Protein Structure. Langmuir 2012, 28, 9673–9679. [Google Scholar] [CrossRef]
- Strojan, K.; Leonardi, A.; Bregar, V.B.; Križaj, I.; Svete, J.; Pavlin, M. Dispersion of Nanoparticles in Different Media Importantly Determines the Composition of Their Protein Corona. PLoS ONE 2017, 12, e0169552. [Google Scholar] [CrossRef] [Green Version]
- Böhmert, L.; Voß, L.; Stock, V.; Braeuning, A.; Lampen, A.; Sieg, H. Isolation Methods for Particle Protein Corona Complexes from Protein-Rich Matrices. Nanoscale Adv. 2020, 2, 563–582. [Google Scholar] [CrossRef] [Green Version]
- Barbir, R.; Goessler, W.; Ćurlin, M.; Micek, V.; Milić, M.; Vuković, B.; Milić, M.; Ljubojević, M.; Jurašin, D.D.; Vrček, I.V. Protein Corona Modulates Distribution and Toxicological Effects of Silver Nanoparticles In Vivo. Part. Part. Syst. Charact. 2019, 36, 1900174. [Google Scholar] [CrossRef]
- Bertrand, N.; Grenier, P.; Mahmoudi, M.; Lima, E.M.; Appel, E.A.; Dormont, F.; Lim, J.-M.; Karnik, R.; Langer, R.; Farokhzad, O.C. Mechanistic Understanding of in Vivo Protein Corona Formation on Polymeric Nanoparticles and Impact on Pharmacokinetics. Nat. Commun. 2017, 8, 777. [Google Scholar] [CrossRef] [PubMed]
- Giri, K.; Kuschnerus, I.; Ruan, J.; Garcia-Bennett, A.E. Influence of a Protein Corona on the Oral Pharmacokinetics of Testosterone Released from Mesoporous Silica. Adv. Ther. 2020, 3, 1900110. [Google Scholar] [CrossRef]
- Tavakoli, S.; Kari, O.K.; Turunen, T.; Lajunen, T.; Schmitt, M.; Lehtinen, J.; Tasaka, F.; Parkkila, P.; Ndika, J.; Viitala, T.; et al. Diffusion and Protein Corona Formation of Lipid-Based Nanoparticles in the Vitreous Humor: Profiling and Pharmacokinetic Considerations. Mol. Pharm. 2020. [Google Scholar] [CrossRef] [PubMed]
- Corbo, C.; Molinaro, R.; Parodi, A.; Toledano Furman, N.E.; Salvatore, F.; Tasciotti, E. The Impact of Nanoparticle Protein Corona on Cytotoxicity, Immunotoxicity and Target Drug Delivery. Nanomedicine 2016, 11, 81–100. [Google Scholar] [CrossRef] [Green Version]
- Mohammad-Beigi, H.; Hayashi, Y.; Zeuthen, C.M.; Eskandari, H.; Scavenius, C.; Juul-Madsen, K.; Vorup-Jensen, T.; Enghild, J.J.; Sutherland, D.S. Mapping and Identification of Soft Corona Proteins at Nanoparticles and Their Impact on Cellular Association. Nat. Commun. 2020, 11, 4535. [Google Scholar] [CrossRef]
- Westmeier, D.; Chen, C.; Stauber, R.H.; Docter, D. The Bio-Corona and Its Impact on Nanomaterial Toxicity. Eur. J.Nanomed. 2015, 7, 153–168. [Google Scholar] [CrossRef]
- Huang, X.; El-Sayed, M.A. Gold Nanoparticles: Optical Properties and Implementations in Cancer Diagnosis and Photothermal Therapy. J. Adv. Res. 2010, 1, 13–28. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Jain, P.K.; El-Sayed, I.H.; El-Sayed, M.A. Gold Nanoparticles: Interesting Optical Properties and Recent Applications in Cancer Diagnostics and Therapy. Nanomedicine 2007, 2, 681–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stetsenko, M.O.; Rudenko, S.P.; Maksimenko, L.S.; Serdega, B.K.; Pluchery, O.; Snegir, S.V. Optical Properties of Gold Nanoparticle Assemblies on a Glass Surface. Nanoscale Res. Lett. 2017, 12, 348. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Bansal, V.; Chaudhary, M.; Basu, A.; Bhonde, R.R.; Sastry, M. Biocompatibility of Gold Nanoparticles and Their Endocytotic Fate inside the Cellular Compartment: A Microscopic Overview. Langmuir 2005, 21, 10644–10654. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, L.; Fan, Y.; Feng, Q.; Cui, F. Biocompatibility and Toxicity of Nanoparticles and Nanotubes. Available online: https://www.hindawi.com/journals/jnm/2012/548389/ (accessed on 4 December 2020).
- Kumar, S.; Jha, I.; Mogha, N.K.; Venkatesu, P. Biocompatibility of Surface-Modified Gold Nanoparticles towards Red Blood Cells and Haemoglobin. Appl. Surf. Sci. 2020, 512, 145573. [Google Scholar] [CrossRef]
- Guo, J.; Rahme, K.; He, Y.; Li, L.-L.; Holmes, J.D.; O’Driscoll, C.M. Gold Nanoparticles Enlighten the Future of Cancer Theranostics. Int. J. Nanomed. 2017, 12, 6131–6152. [Google Scholar] [CrossRef] [Green Version]
- Anderson, L.J.E.; Payne, C.M.; Zhen, Y.-R.; Nordlander, P.; Hafner, J.H. A Tunable Plasmon Resonance in Gold Nanobelts. Nano Lett. 2011, 11, 5034–5037. [Google Scholar] [CrossRef]
- Lal, S.; Hafner, J.H.; Halas, N.J.; Link, S.; Nordlander, P. Noble Metal Nanowires: From Plasmon Waveguides to Passive and Active Devices. Acc. Chem. Res. 2012, 45, 1887–1895. [Google Scholar] [CrossRef]
- Nehl, C.L.; Liao, H.; Hafner, J.H. Optical Properties of Star-Shaped Gold Nanoparticles. Nano Lett. 2006, 6, 683–688. [Google Scholar] [CrossRef] [Green Version]
- Brinson, B.E.; Lassiter, J.B.; Levin, C.S.; Bardhan, R.; Mirin, N.; Halas, N.J. Nanoshells Made Easy: Improving Au Layer Growth on Nanoparticle Surfaces. Langmuir 2008, 24, 14166–14171. [Google Scholar] [CrossRef] [Green Version]
- Kelly, A.T.; Filgueira, C.S.; Schipper, D.E.; Halas, N.J.; Whitmire, K.H. Gold Coated Iron Phosphide Core–Shell Structures. RSC Adv. 2017, 7, 25848–25854. [Google Scholar] [CrossRef] [Green Version]
- Levin, C.S.; Hofmann, C.; Ali, T.A.; Kelly, A.T.; Morosan, E.; Nordlander, P.; Whitmire, K.H.; Halas, N.J. Magnetic-Plasmonic Core-Shell Nanoparticles. ACS Nano 2009, 3, 1379–1388. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.-R.; Stanton-Maxey, K.J.; Stanley, J.K.; Levin, C.S.; Bardhan, R.; Akin, D.; Badve, S.; Sturgis, J.; Robinson, J.P.; Bashir, R.; et al. A Cellular Trojan Horse for Delivery of Therapeutic Nanoparticles into Tumors. Nano Lett. 2007, 7, 3759–3765. [Google Scholar] [CrossRef]
- Rastinehad, A.R.; Anastos, H.; Wajswol, E.; Winoker, J.S.; Sfakianos, J.P.; Doppalapudi, S.K.; Carrick, M.R.; Knauer, C.J.; Taouli, B.; Lewis, S.C.; et al. Gold Nanoshell-Localized Photothermal Ablation of Prostate Tumors in a Clinical Pilot Device Study. Proc. Natl. Acad. Sci. USA 2019, 116, 18590–18596. [Google Scholar] [CrossRef] [Green Version]
- Bros, M.; Nuhn, L.; Simon, J.; Moll, L.; Mailänder, V.; Landfester, K.; Grabbe, S. The Protein Corona as a Confounding Variable of Nanoparticle-Mediated Targeted Vaccine Delivery. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Brun, E.; Sicard-Roselli, C. Could Nanoparticle Corona Characterization Help for Biological Consequence Prediction? Cancer Nanotechnol. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Hong, J.; Ding, Y. Biological Behavior Regulation of Gold Nanoparticles via the Protein Corona. Adv. Healthc. Mater. 2020, 9, e1901448. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ganesh, S.; Wang, W.; Amiji, M. Protein Corona-Enabled Systemic Delivery and Targeting of Nanoparticles. AAPS J. 2020, 22, 83. [Google Scholar] [CrossRef] [PubMed]
- Piella, J.; Bastús, N.G.; Puntes, V. Size-Dependent Protein–Nanoparticle Interactions in Citrate-Stabilized Gold Nanoparticles: The Emergence of the Protein Corona. Bioconj. Chem. 2017, 28, 88–97. [Google Scholar] [CrossRef]
- Li, B.; Lane, L.A. Probing the Biological Obstacles of Nanomedicine with Gold Nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019, 11. [Google Scholar] [CrossRef]
- Mosquera, J.; García, I.; Henriksen-Lacey, M.; Martínez-Calvo, M.; Dhanjani, M.; Mascareñas, J.L.; Liz-Marzán, L.M. Reversible Control of Protein Corona Formation on Gold Nanoparticles Using Host–Guest Interactions. ACS Nano 2020, 14, 5382–5391. [Google Scholar] [CrossRef] [PubMed]
- Elechalawar, C.K.; Hossen, M.N.; McNally, L.; Bhattacharya, R.; Mukherjee, P. Analysing the Nanoparticle-Protein Corona for Potential Molecular Target Identification. J. Control. Release 2020, 322, 122–136. [Google Scholar] [CrossRef]
- Park, S.J. Protein–Nanoparticle Interaction: Corona Formation and Conformational Changes in Proteins on Nanoparticles. Int. J. Nanomed. 2020, 15, 5783–5802. [Google Scholar] [CrossRef] [PubMed]
- Nierenberg, D.; Khaled, A.R.; Flores, O. Formation of a Protein Corona Influences the Biological Identity of Nanomaterials. Rep. Pract. Oncol. Radiother. 2018, 23, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Fehér, B.; Kitka, D.; Wacha, A.; Bóta, A.; Berényi, S.; Pipich, V.; Fraikin, J.-L. Size Measurement of Extracellular Vesicles and Synthetic Liposomes: The Impact of the Hydration Shell and the Protein Corona. Colloids Surf. B Biointerfaces 2020, 192, 111053. [Google Scholar] [CrossRef] [PubMed]
- Digiacomo, L.; Palchetti, S.; Giulimondi, F.; Pozzi, D.; Chiozzi, R.Z.; Capriotti, A.L.; Laganà, A.; Caracciolo, G. The Biomolecular Corona of Gold Nanoparticles in a Controlled Microfluidic Environment. Lab. Chip. 2019, 19, 2557–2567. [Google Scholar] [CrossRef]
- Cheng, X.; Tian, X.; Wu, A.; Li, J.; Tian, J.; Chong, Y.; Chai, Z.; Zhao, Y.; Chen, C.; Ge, C. Protein Corona Influences Cellular Uptake of Gold Nanoparticles by Phagocytic and Nonphagocytic Cells in a Size-Dependent Manner. ACS Appl. Mater. Interfaces 2015, 7, 20568–20575. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of Nanoparticle Design for Overcoming Biological Barriers to Drug Delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Chandran, P.; Riviere, J.E.; Monteiro-Riviere, N.A. Surface Chemistry of Gold Nanoparticles Determines the Biocorona Composition Impacting Cellular Uptake, Toxicity and Gene Expression Profiles in Human Endothelial Cells. Nanotoxicology 2017, 11, 507–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.; Zhu, L.; Mo, J.; Wei, W.; Yuan, B.; Zhao, J.; Cao, C. Protein Corona and Immune Responses of Borophene: A Comparison of Nanosheet–Plasma Interface with Graphene and Phosphorene. ACS Appl. Bio Mater. 2020, 3, 4220–4229. [Google Scholar] [CrossRef]
- Neagu, M.; Piperigkou, Z.; Karamanou, K.; Engin, A.B.; Docea, A.O.; Constantin, C.; Negrei, C.; Nikitovic, D.; Tsatsakis, A. Protein Bio-Corona: Critical Issue in Immune Nanotoxicology. Arch. Toxicol. 2017, 91, 1031–1048. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.Y.; Kim, H.S.; Palanikumar, L.; Go, E.M.; Jana, B.; Park, S.A.; Kim, H.Y.; Kim, K.; Seo, J.K.; Kwak, S.K.; et al. Cloaking Nanoparticles with Protein Corona Shield for Targeted Drug Delivery. Nat. Commun. 2018, 9, 4548. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; He, X.; Zhang, Z.; Zhao, Y.; Feng, W. Metabolism of Nanomaterials in Vivo: Blood Circulation and Organ Clearance. Acc. Chem. Res. 2013, 46, 761–769. [Google Scholar] [CrossRef]
- Gullino, P.M.; Clark, S.H.; Grantham, F.H. The Interstitial Fluid of Solid Tumors. Cancer Res. 1964, 24, 780–797. [Google Scholar]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.-K. Non-Small-Cell Lung Cancers: A Heterogeneous Set of Diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef]
- Van Lamsweerde, A.L.; Henry, N.; Vaes, G. Metastatic Heterogeneity of Cells from Lewis Lung Carcinoma. Cancer Res. 1983, 43, 5314–5320. [Google Scholar]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Vighetto, V.; Di Marzio, N.; Ferraro, F.; Hirsch, M.; Ferrante, N.; Mitra, S.; Grattoni, A.; Filgueira, C.S. Gold Nanoparticles Radio-Sensitize and Reduce Cell Survival in Lewis Lung Carcinoma. Nanomaterials 2020, 10, 1717. [Google Scholar] [CrossRef]
- Terracciano, R.; Sprouse, M.L.; Wang, D.; Ricchetti, S.; Hirsch, M.; Ferrante, N.; Butler, E.B.; Demarchi, D.; Grattoni, A.; Filgueira, C.S. Intratumoral Gold Nanoparticle-Enhanced CT Imaging: An in Vivo Investigation of Biodistribution and Retention. In Proceedings of the IEEE 20th International Conference on Nanotechnology, Montreal, QC, Canada, 28–31 July 2020; pp. 349–353. [Google Scholar] [CrossRef]
- Hughes, H.J.; Demers, S.M.E.; Zhang, A.; Hafner, J.H. The Orientation of a Membrane Probe from Structural Analysis by Enhanced Raman Scattering. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183109. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef] [Green Version]
- Baimanov, D.; Cai, R.; Chen, C. Understanding the Chemical Nature of Nanoparticle–Protein Interactions. Bioconjugate Chem. 2019, 30, 1923–1937. [Google Scholar] [CrossRef] [PubMed]
- Sotnikov, D.V.; Berlina, A.N.; Ivanov, V.S.; Zherdev, A.V.; Dzantiev, B.B. Adsorption of Proteins on Gold Nanoparticles: One or More Layers? Colloids Surf. B Biointerfaces 2019, 173, 557–563. [Google Scholar] [CrossRef]
- Jommanee, N.; Chanthad, C.; Manokruang, K. Preparation of Injectable Hydrogels from Temperature and PH Responsive Grafted Chitosan with Tuned Gelation Temperature Suitable for Tumor Acidic Environment. Carbohydr. Polym. 2018, 198, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Medina, S.; McDonough, S.; Swanglap, P.; Landes, C.F.; Link, S. In Situ Measurement of Bovine Serum Albumin Interaction with Gold Nanospheres. Langmuir 2012, 28, 9131–9139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Röcker, C.; Pötzl, M.; Zhang, F.; Parak, W.J.; Nienhaus, G.U. A Quantitative Fluorescence Study of Protein Monolayer Formation on Colloidal Nanoparticles. Nat. Nanotech. 2009, 4, 577–580. [Google Scholar] [CrossRef]
- Campeau, E.; Ruhl, V.E.; Rodier, F.; Smith, C.L.; Rahmberg, B.L.; Fuss, J.O.; Campisi, J.; Yaswen, P.; Cooper, P.K.; Kaufman, P.D. A Versatile Viral System for Expression and Depletion of Proteins in Mammalian Cells. PLoS ONE 2009, 4, e6529. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.C.; Hajfathalian, M.; Maidment, P.S.N.; Hsu, J.C.; Naha, P.C.; Si-Mohamed, S.; Breuilly, M.; Kim, J.; Chhour, P.; Douek, P.; et al. Effect of Gold Nanoparticle Size on Their Properties as Contrast Agents for Computed Tomography. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Szekeres, G.P.; Kneipp, J. SERS Probing of Proteins in Gold Nanoparticle Agglomerates. Front. Chem. 2019, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, H.; Kushwaha, A.; Kumar, A.; Aslam, M. A Facile PH Controlled Citrate-Based Reduction Method for Gold Nanoparticle Synthesis at Room Temperature. Nanoscale Res. Lett. 2016, 11, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, S.; Kojio, K.; Takahara, A.; Kajiyama, T. Bovine Serum Albumin Adsorption onto Immobilized Organotrichlorosilane Surface: Influence of the Phase Separation on Protein Adsorption Patterns. J. Biomater. Sci. Polym. Ed. 1998, 9, 131–150. [Google Scholar] [CrossRef]
- Sutoo, S.; Maeda, T.; Suzuki, A.; Kato, Y. Adaptation to Chronic Acidic Extracellular PH Elicits a Sustained Increase in Lung Cancer Cell Invasion and Metastasis. Clin. Exp. Metastasis 2020, 37, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Oh, N.; Park, J.-H. Endocytosis and Exocytosis of Nanoparticles in Mammalian Cells. Int. J. Nanomed. 2014, 9, 51–63. [Google Scholar] [CrossRef] [Green Version]
- Mocan, L.; Matea, C.; Tabaran, F.A.; Mosteanu, O.; Pop, T.; Puia, C.; Agoston-Coldea, L.; Zaharie, G.; Mocan, T.; Buzoianu, A.D.; et al. Selective Ex Vivo Photothermal Nano-Therapy of Solid Liver Tumors Mediated by Albumin Conjugated Gold Nanoparticles. Biomaterials 2017, 119, 33–42. [Google Scholar] [CrossRef]
- Charbgoo, F.; Nejabat, M.; Abnous, K.; Soltani, F.; Taghdisi, S.M.; Alibolandi, M.; Thomas Shier, W.; Steele, T.W.J.; Ramezani, M. Gold Nanoparticle Should Understand Protein Corona for Being a Clinical Nanomaterial. J. Control. Release 2018, 272, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Rampado, R.; Crotti, S.; Caliceti, P.; Pucciarelli, S.; Agostini, M. Recent Advances in Understanding the Protein Corona of Nanoparticles and in the Formulation of “Stealthy” Nanomaterials. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Rosa, S.; Connolly, C.; Schettino, G.; Butterworth, K.T.; Prise, K.M. Biological Mechanisms of Gold Nanoparticle Radiosensitization. Cancer Nanotechnol. 2017, 8, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terracciano, R.; Zhang, A.; Butler, E.B.; Demarchi, D.; Hafner, J.H.; Grattoni, A.; Filgueira, C.S. Effects of Surface Protein Adsorption on the Distribution and Retention of Intratumorally Administered Gold Nanoparticles. Pharmaceutics 2021, 13, 216. https://doi.org/10.3390/pharmaceutics13020216
Terracciano R, Zhang A, Butler EB, Demarchi D, Hafner JH, Grattoni A, Filgueira CS. Effects of Surface Protein Adsorption on the Distribution and Retention of Intratumorally Administered Gold Nanoparticles. Pharmaceutics. 2021; 13(2):216. https://doi.org/10.3390/pharmaceutics13020216
Chicago/Turabian StyleTerracciano, Rossana, Aobo Zhang, E. Brian Butler, Danilo Demarchi, Jason H. Hafner, Alessandro Grattoni, and Carly S. Filgueira. 2021. "Effects of Surface Protein Adsorption on the Distribution and Retention of Intratumorally Administered Gold Nanoparticles" Pharmaceutics 13, no. 2: 216. https://doi.org/10.3390/pharmaceutics13020216