Development of Novel Peptides for the Antimicrobial Combination Therapy against Carbapenem-Resistant Acinetobacter baumannii Infection
Abstract
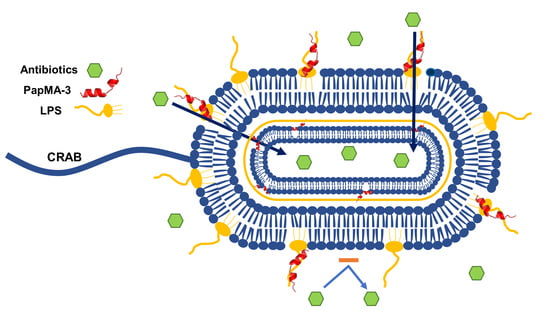
:1. Introduction
2. Materials and Methods
2.1. Peptides and Materials
2.2. Antimicrobial Activity
2.3. Peptide-LPS Binding Assay
2.4. Membrane Depolarization
2.5. Time-Dependent Permeabilization of the Outer Membrane
2.6. Cell Cytotoxicity
2.7. Stability of PapMA-3 Compared to Melittin in Human Serum
2.8. Hemolytic Activity
2.9. Checkerboard Assays
2.10. Time Killing Assay
2.11. Scanning Electron Microscope Analysis
2.12. Biofilm Inhibition
2.13. Isothermal Titration Calorimetry (ITC)
2.14. Saturation Transfer Difference (STD)-NMR
2.15. Statistical Analysis
3. Results
3.1. Design of PapMA and Its Analogs
| Peptides | Sequence | Length | Molecular Weight | Hydrophobicity <H> 1 | Net Charge 2 |
|---|---|---|---|---|---|
| papiliocin | RWKIFKKIEKVGRNVRDGIIKAGPAVAVVGQAATVVK-NH2 | 37 | 4002.8 | 0.300 | 7 |
| Magainin 2 | GIGKFLHSAKKFGKAFVGEIMNS | 23 | 2466.9 | 0.373 | 3 |
| PapMA | RWKIFKKIPKFLHSAKKF-NH2 | 18 | 2302.1 | 0.394 | 7 |
| PapMA-2 | RWKIFKKIPKFLHSAKKA-NH2 | 18 | 2225.5 | 0.312 | 7 |
| PapMA-3 | RWKIFKKIPKFLHSAKKW-NH2 | 18 | 2340.6 | 0.419 | 7 |
| PapMA-4 | RWKIFKKIPKFLHSFKKF-NH2 | 18 | 2377.5 | 0.476 | 7 |
| PapMA-5 | RWKIFKKIPKFLHSWKKF-NH2 | 18 | 2416.4 | 0.502 | 7 |
| PapMA-6 | RWKIFKKIPKFLHSWKKW-NH2 | 18 | 2455.6 | 0.527 | 7 |
3.2. Antimicrobial Activities of PapMA Analogs
3.3. Antibacterial Mechanisms of PapMA Analogs against CRAB
3.3.1. Binding Assay of LPS-PapMA Analogs
3.3.2. Depolarization of PapMA and Its Analogs against CRAB C1
3.4. Cytotoxicities of PapMA Analogs
3.5. Synergistic Effects of PapMA-3 with Antibiotics against Five CRAB
3.6. Mechanism of Synergistic Activity of PapMA-3 with Antibiotics against CRAB
3.6.1. Confirmation of Synergistic Effects between PapMA-3 and Antibiotics through Time Killing Assays
3.6.2. Visualization of CRAB C1 Membrane Disruption by PapMA-3 in Combination with Antibiotics Using a Field Emission Scanning Electron Microscope (FE-SEM)
3.7. Synergistic Effects of PapMA-3 on Biofilm Inhibition
3.8. Stability and Effects of PapMA-3 on Mammalian Cells Compared to That of Melittin
3.8.1. Stability of PapMA-3 Compared to That of Melittin in the Presence of Human Serum
3.8.2. Effects of PapMA-3 Compared to That of Melittin on Mammalian Cells
3.9. Binding Interactions of PapMA-3 with LPS as Studied by STD-NMR Spectroscopy and ITC
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Neu, H.C. Infections due to gram-negative bacteria: An overview. Rev. Infect. Dis. 1985, 7 (Suppl. 4), S778–S782. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Vasoo, S.; Barreto, J.N.; Tosh, P.K. Emerging issues in gram-negative bacterial resistance: An update for the practicing clinician. Mayo Clin. Proc. 2015, 90, 395–403. [Google Scholar] [CrossRef] [Green Version]
- Deelen, J.W.T.; Rottier, W.C.; van Werkhoven, C.H.; Woudt, S.H.S.; Buiting, A.G.M.; Dorigo-Zetsma, J.W.; Kluytmans, J.; van der Linden, P.D.; Thijsen, S.F.T.; Vlaminckx, B.J.M.; et al. The burden of bacteremic and non-bacteremic Gram-negative infections: A prospective multicenter cohort study in a low-resistance country. J. Infect. 2020, 81, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Lien, C.Y.; Lee, J.J.; Tsai, W.C.; Chen, S.Y.; Huang, C.R.; Chien, C.C.; Lu, C.H.; Chang, W.N. The clinical characteristics of spontaneous Gram-negative bacterial meningitis in adults: A hospital-based study. J. Clin. Neurosci. 2019, 64, 101–105. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in Health Care Facilities; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Shrivastava, S.R.; Shrivastava, P.S.; Ramasamy, J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med Soc. 2018, 32, 76. [Google Scholar] [CrossRef]
- Sleiman, A.; Fayad, A.G.A.; Banna, H.; Matar, G.M. Prevalence and molecular epidemiology of carbapenem-resistant Gram-negative bacilli and their resistance determinants in the Eastern Mediterranean Region over the last decade. J. Glob. Antimicrob. Resist. 2021, 25, 209–221. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Patocka, J.; Kuca, K. Insect Antimicrobial Peptides, a Mini Review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Diamond, G.; Beckloff, N.; Weinberg, A.; Kisich, K.O. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 2009, 15, 2377–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Narayana, J.L.; Chen, J.Y. Antimicrobial peptides: Possible anti-infective agents. Peptides 2015, 72, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2011, 11, 37–51. [Google Scholar] [CrossRef]
- Datta, S.; Roy, A. Antimicrobial Peptides as Potential Therapeutic Agents: A Review. Int. J. Pept. Res. Ther. 2021, 27, 555–577. [Google Scholar] [CrossRef]
- Kim, M.K.; Lee, T.G.; Jung, M.; Park, K.H.; Chong, Y. In Vitro Synergism and Anti-biofilm Activity of Quercetin-Pivaloxymethyl Conjugate against Staphylococcus aureus and Enterococcus Species. Chem. Pharm. Bull. 2018, 66, 1019–1022. [Google Scholar] [CrossRef]
- Pachon-Ibanez, M.E.; Docobo-Perez, F.; Lopez-Rojas, R.; Dominguez-Herrera, J.; Jimenez-Mejias, M.E.; Garcia-Curiel, A.; Pichardo, C.; Jimenez, L.; Pachon, J. Efficacy of rifampin and its combinations with imipenem, sulbactam, and colistin in experimental models of infection caused by imipenem-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 1165–1172. [Google Scholar] [CrossRef] [Green Version]
- Mgbeahuruike, E.E.; Stalnacke, M.; Vuorela, H.; Holm, Y. Antimicrobial and Synergistic Effects of Commercial Piperine and Piperlongumine in Combination with Conventional Antimicrobials. Antibiotics 2019, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Assane, I.M.; Santos-Filho, N.A.; de Sousa, E.L.; de Arruda Brasil, M.C.O.; Cilli, E.M.; Pilarski, F. Cytotoxicity and antimicrobial activity of synthetic peptides alone or in combination with conventional antimicrobials against fish pathogenic bacteria. J. Appl. Microbiol. 2021, 131, 1762–1774. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, H.; Alikhani, M.Y.; Imani Fooladi, A.A. Synergistic antimicrobial activity of melittin with clindamycin on the expression of encoding exfoliative toxin in Staphylococcus aureus. Toxicon 2020, 183, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Rishi, P.; Vij, S.; Maurya, I.K.; Kaur, U.J.; Bharati, S.; Tewari, R. Peptides as adjuvants for ampicillin and oxacillin against methicillin-resistant Staphylococcus aureus (MRSA). Microb. Pathog. 2018, 124, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Chen, C.; Wang, D.; Tong, Z.; Wang, Z.; Liu, Y. Amphipathic Peptide Antibiotics with Potent Activity against Multidrug-Resistant Pathogens. Pharmaceutics 2021, 13, 438. [Google Scholar] [CrossRef] [PubMed]
- Gan, B.H.; Cai, X.; Javor, S.; Kohler, T.; Reymond, J.L. Synergistic Effect of Propidium Iodide and Small Molecule Antibiotics with the Antimicrobial Peptide Dendrimer G3KL against Gram-Negative Bacteria. Molecules 2020, 25, 5643. [Google Scholar] [CrossRef] [PubMed]
- Pollini, S.; Brunetti, J.; Sennati, S.; Rossolini, G.M.; Bracci, L.; Pini, A.; Falciani, C. Synergistic activity profile of an antimicrobial peptide against multidrug-resistant and extensively drug-resistant strains of Gram-negative bacterial pathogens. J. Pept. Sci. 2017, 23, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Akbari, R.; Hakemi-Vala, M.; Pashaie, F.; Bevalian, P.; Hashemi, A.; Pooshang Bagheri, K. Highly Synergistic Effects of Melittin with Conventional Antibiotics Against Multidrug-Resistant Isolates of Acinetobacter baumannii and Pseudomonas aeruginosa. Microb. Drug Resist. 2019, 25, 193–202. [Google Scholar] [CrossRef]
- Shin, S.Y.; Kang, J.H.; Lee, M.K.; Kim, S.Y.; Kim, Y.; Hahm, K.S. Cecropin A—magainin 2 hybrid peptides having potent antimicrobial activity with low hemolytic effect. Biochem. Mol. Biol. Int. 1998, 44, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.Y.; Lee, M.K.; Kim, K.L.; Hahm, K.S. Structure-antitumor and hemolytic activity relationships of synthetic peptides derived from cecropin A-magainin 2 and cecropin A-melittin hybrid peptides. J. Pept. Res. 1997, 50, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Shin, S.Y.; Kang, J.H.; Hahm, K.S.; Kim, K.L.; Kim, Y. NMR structural characterization of cecropin A(1-8)—magainin 2(1-12) and cecropin A (1-8)—melittin (1-12) hybrid peptides. J. Pept. Res. 1999, 53, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Shin, A.; Lee, E.; Jeon, D.; Park, Y.G.; Bang, J.K.; Park, Y.S.; Shin, S.Y.; Kim, Y. Peptoid-Substituted Hybrid Antimicrobial Peptide Derived from Papiliocin and Magainin 2 with Enhanced Bacterial Selectivity and Anti-inflammatory Activity. Biochemistry 2015, 54, 3921–3931. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, M.; Choi, J.; Jang, A.; Kim, Y. A Novel Peptide Antibiotic, Pro10-1D, Designed from Insect Defensin Shows Antibacterial and Anti-Inflammatory Activities in Sepsis Models. Int. J. Mol. Sci. 2020, 21, 6216. [Google Scholar] [CrossRef]
- Krishnan, M.; Choi, J.; Choi, S.; Kim, Y. Anti-Endotoxin 9-Meric Peptide with Therapeutic Potential for the Treatment of Endotoxemia. J. Microbiol. Biotechnol. 2021, 31, 25–32. [Google Scholar] [CrossRef]
- Golec, M. Cathelicidin LL-37: LPS-neutralizing, pleiotropic peptide. Ann. Agric. Environ. Med. 2007, 14, 1–4. [Google Scholar] [PubMed]
- Jeon, D.; Jacob, B.; Kwak, C.; Kim, Y. Short Antimicrobial Peptides Exhibiting Antibacterial and Anti-Inflammatory Activities Derived from the N-Terminal Helix of Papiliocin. Bull. Korean Chem. Soc. 2017, 38, 1260–1268. [Google Scholar] [CrossRef]
- Lee, E.; Kim, J.K.; Jeon, D.; Jeong, K.W.; Shin, A.; Kim, Y. Functional Roles of Aromatic Residues and Helices of Papiliocin in its Antimicrobial and Anti-inflammatory Activities. Sci. Rep. 2015, 5, 12048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonapace, C.R.; Bosso, J.A.; Friedrich, L.V.; White, R.L. Comparison of methods of interpretation of checkerboard synergy testing. Diagn. Microbiol. Infect. Dis. 2002, 44, 363–366. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Terminology relating to methods for the determination of susceptibility of bacteria to antimicrobial agents. Clin. Microbiol. Infect. 2000, 6, 503–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.D.; Shin, S.Y. Antimicrobial and anti-inflammatory activities of short dodecapeptides derived from duck cathelicidin: Plausible mechanism of bactericidal action and endotoxin neutralization. Eur. J. Med. Chem. 2020, 204, 112580. [Google Scholar] [CrossRef]
- Kim, J.; Jacob, B.; Jang, M.; Kwak, C.; Lee, Y.; Son, K.; Lee, S.; Jung, I.D.; Jeong, M.S.; Kwon, S.H.; et al. Development of a novel short 12-meric papiliocin-derived peptide that is effective against Gram-negative sepsis. Sci. Rep. 2019, 9, 3817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, M.; Kim, J.; Choi, Y.; Bang, J.; Kim, Y. Antiseptic Effect of Ps-K18: Mechanism of Its Antibacterial and Anti-Inflammatory Activities. Int. J. Mol. Sci. 2019, 20, 4895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic alpha helical antimicrobial peptides. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Hong, M.Y.; Park, S.W.; Choi, K.H.; Yun, E.Y.; Goo, T.W.; Kang, S.W.; Suh, H.J.; Kim, I.; Hwang, J.S. Characterization and cDNA cloning of a cecropin-like antimicrobial peptide, papiliocin, from the swallowtail butterfly, Papilio xuthus. Mol. Cells 2010, 29, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 1987, 84, 5449–5453. [Google Scholar] [CrossRef] [Green Version]
- Maloy, W.L.; Kari, U.P. Structure-activity studies on magainins and other host defense peptides. Biopolymers 1995, 37, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Simor, A.E.; Vercaigne, L.; Mandell, L.; Canadian Carbapenem Discussion Group. Imipenem and meropenem: Comparison of in vitro activity, pharmacokinetics, clinical trials and adverse effects. Can. J. Infect. Dis. 1998, 9, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S.A. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell 2001, 104, 901–912. [Google Scholar] [CrossRef]
- Nagarajan, R. Antibacterial activities and modes of action of vancomycin and related glycopeptides. Antimicrob. Agents Chemother. 1991, 35, 605–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papich, M.G. Saunders Handbook of Veterinary Drugs: Small and Large Animal, 4th ed.; Elsevier: St. Louis, MO, USA, 2016; p. 24. 900p. [Google Scholar]
- Mao, J.C.; Putterman, M. Accumulation in gram-postive and gram-negative bacteria as a mechanism of resistance to erythromycin. J. Bacteriol. 1968, 95, 1111–1117. [Google Scholar] [CrossRef] [Green Version]
- Watanakunakorn, C. Mode of action and in-vitro activity of vancomycin. J. Antimicrob. Chemother. 1984, 14 (Suppl. D), 7–18. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, D.; Pizzirani, C.; Adinolfi, E.; Forchap, S.; Sitta, B.; Turchet, L.; Falzoni, S.; Minelli, M.; Baricordi, R.; Di Virgilio, F. The antibiotic polymyxin B modulates P2X7 receptor function. J. Immunol. 2004, 173, 4652–4660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nation, R.L.; Rigatto, M.H.P.; Falci, D.R.; Zavascki, A.P. Polymyxin Acute Kidney Injury: Dosing and Other Strategies to Reduce Toxicity. Antibiotics 2019, 8, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nageeb, W.; Metwally, L.; Kamel, M.; Zakaria, S. In vitro antimicrobial synergy studies of carbapenem-resistant Acinetobacter baumannii isolated from intensive care units of a tertiary care hospital in Egypt. J. Infect. Public Health 2015, 8, 593–602. [Google Scholar] [CrossRef] [Green Version]
- Nagarajan, D.; Roy, N.; Kulkarni, O.; Nanajkar, N.; Datey, A.; Ravichandran, S.; Thakur, C.; Sandeep, T.; Aprameya, I.V.; Sarma, S.P.; et al. Omega76: A designed antimicrobial peptide to combat carbapenem- and tigecycline-resistant Acinetobacter baumannii. Sci. Adv. 2019, 5, eaax1946. [Google Scholar] [CrossRef] [Green Version]
- Bozkurt-Guzel, C.; Inci, G.; Oyardi, O.; Savage, P.B. Synergistic Activity of Ceragenins Against Carbapenem-Resistant Acinetobacter baumannii Strains in Both Checkerboard and Dynamic Time-Kill Assays. Curr. Microbiol. 2020, 77, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- Bonapace, C.R.; White, R.L.; Friedrich, L.V.; Bosso, J.A. Evaluation of antibiotic synergy against Acinetobacter baumannii: A comparison with Etest, time-kill, and checkerboard methods. Diagn. Microbiol. Infect. Dis. 2000, 38, 43–50. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Shamiri, M.M.; Zhang, S.; Mi, P.; Liu, Y.; Xun, M.; Yang, E.; Ai, L.; Han, L.; Chen, Y. Phenotypic and genotypic characteristics of Acinetobacter baumannii enrolled in the relationship among antibiotic resistance, biofilm formation and motility. Microb. Pathog. 2021, 155, 104922. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, M.; Novak-Frazer, L.; Bates, M.; Baguneid, M.; Alonso-Rasgado, T.; Xia, G.; Rautemaa-Richardson, R.; Bayat, A. Validation of biofilm formation on human skin wound models and demonstration of clinically translatable bacteria-specific volatile signatures. Sci. Rep. 2018, 8, 9431. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.; Benoliel, C.; Drider, D.; Dhulster, P.; Chihib, N.E. Biofilm formation and persistence on abiotic surfaces in the context of food and medical environments. Arch. Microbiol. 2014, 196, 453–472. [Google Scholar] [CrossRef] [PubMed]
- Longo, F.; Vuotto, C.; Donelli, G. Biofilm formation in Acinetobacter baumannii. New Microbiol. 2014, 37, 119–127. [Google Scholar]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, E.; Shin, S.; Jeong, K.W.; Lee, J.Y.; Bae, S.Y.; Kim, S.H.; Lee, J.; Kim, S.R.; Lee, D.G.; et al. Structure and function of papiliocin with antimicrobial and anti-inflammatory activities isolated from the swallowtail butterfly, Papilio xuthus. J. Biol. Chem. 2011, 286, 41296–41311. [Google Scholar] [CrossRef] [Green Version]
- Wright, H.; Bonomo, R.A.; Paterson, D.L. New agents for the treatment of infections with Gram-negative bacteria: Restoring the miracle or false dawn? Clin. Microbiol. Infect. 2017, 23, 704–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piperaki, E.T.; Tzouvelekis, L.S.; Miriagou, V.; Daikos, G.L. Carbapenem-resistant Acinetobacter baumannii: In pursuit of an effective treatment. Clin. Microbiol. Infect. 2019, 25, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Viehman, J.A.; Nguyen, M.H.; Doi, Y. Treatment options for carbapenem-resistant and extensively drug-resistant Acinetobacter baumannii infections. Drugs 2014, 74, 1315–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenhard, J.R.; Nation, R.L.; Tsuji, B.T. Synergistic combinations of polymyxins. Int. J. Antimicrob. Agents 2016, 48, 607–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.J.; Cho, J.H.; Kim, H.J.; Han, S.H.; Jeong, S.H.; Byun, M.K. Colistin monotherapy versus colistin/rifampicin combination therapy in pneumonia caused by colistin-resistant Acinetobacter baumannii: A randomised controlled trial. J. Glob. Antimicrob. Resist. 2019, 17, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Nigro, S.J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genomics 2019, 5, e000306. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.H.; Chan, B.K.; Chan, E.W.; Chen, S. Over-Expression of ISAba1-Linked Intrinsic and Exogenously Acquired OXA Type Carbapenem-Hydrolyzing-Class D-ss-Lactamase-Encoding Genes Is Key Mechanism Underlying Carbapenem Resistance in Acinetobacter baumannii. Front. Microbiol. 2019, 10, 2809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pristovsek, P.; Kidric, J. Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: An NMR and molecular modeling study. J. Med. Chem. 1999, 42, 4604–4613. [Google Scholar] [CrossRef]
- Mares, J.; Kumaran, S.; Gobbo, M.; Zerbe, O. Interactions of lipopolysaccharide and polymyxin studied by NMR spectroscopy. J. Biol. Chem. 2009, 284, 11498–11506. [Google Scholar] [CrossRef] [Green Version]
- Sahoo, A.; Swain, S.S.; Behera, A.; Sahoo, G.; Mahapatra, P.K.; Panda, S.K. Antimicrobial Peptides Derived From Insects Offer a Novel Therapeutic Option to Combat Biofilm: A Review. Front. Microbiol. 2021, 12, 661195. [Google Scholar] [CrossRef] [PubMed]
- Kapil, S.; Sharma, V. d-Amino acids in antimicrobial peptides: A potential approach to treat and combat antimicrobial resistance. Can. J. Microbiol. 2021, 67, 119–137. [Google Scholar] [CrossRef] [PubMed]
- Hicks, R.P.; Abercrombie, J.J.; Wong, R.K.; Leung, K.P. Antimicrobial peptides containing unnatural amino acid exhibit potent bactericidal activity against ESKAPE pathogens. Bioorg. Med. Chem. 2013, 21, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Falanga, A.; Nigro, E.; De Biasi, M.G.; Daniele, A.; Morelli, G.; Galdiero, S.; Scudiero, O. Cyclic Peptides as Novel Therapeutic Microbicides: Engineering of Human Defensin Mimetics. Molecules 2017, 22, 1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]











| Peptides | Minimum Inhibitory Concentration (μg·mL−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gram-Negative Bacteria | Gram-Positive Bacteria | ||||||||
| E. coli | A. baumanii | CRAB C1 | CRAB C2 | CRAB C3 | CRAB C4 | CRAB C5 | GM 1 | S.aureus | |
| PapMA | 32 | 32 | 32 | 32 | 16 | 64 | 32 | 34 | 64 |
| PapMA-2 | 64 | 128 | 128 | >128 | 64 | >128 | 128 | 146 | >128 |
| PapMA-3 | 16 | 16 | 16 | 16 | 16 | 32 | 16 | 18 | 32 |
| PapMA-4 | 16 | 8 | 8 | 8 | 8 | 16 | 8 | 10 | 32 |
| PapMA-5 | 16 | 16 | 16 | 16 | 16 | 32 | 16 | 18 | 32 |
| PapMA-6 | 16 | 8 | 8 | 8 | 8 | 16 | 16 | 11 | 32 |
| melittin | 8 | 16 | 16 | 16 | 16 | 8 | 16 | 14 | 16 |
| Antibiotics | |||||||||
| Imipenem | 0.25 | 0.25 | 64 | 64 | 64 | 64 | 64 | 46 | 1 |
| Meropenem | 0.25 | 0.25 | 128 | 64 | 64 | 128 | 64 | 64 | 1 |
| Rifampin | 2 | 4 | 128 | 64 | 128 | 128 | 256 | 101 | 1 |
| Erythromycin | 16 | 32 | >512 | 512 | 512 | >512 | >512 | 592 | 0.25 |
| Vancomycin | 128 | 256 | 256 | 256 | 256 | 512 | 256 | 274 | 0.5 |
| Linezolid | 256 | 256 | 256 | 256 | 256 | 512 | 512 | 329 | 1 |
| PapMA-3 with Antibiotics | MIC (μg·mL−1) | MIC in Combination (μg·mL−1) | ||||
|---|---|---|---|---|---|---|
| Strain | PapMA-3 | Antibiotics | PapMA-3 | Antibiotics | FICI # | |
| PapMA-3 + Imipenem | CRAB C1 | 16 | 64 | 4.0 | 4.0 | 0.31 * |
| CRAB C2 | 16 | 64 | 4.0 | 8.0 | 0.38 | |
| CRAB C3 | 16 | 64 | 8.0 | 1.0 | 0.52 | |
| CRAB C4 | 32 | 64 | 8.0 | 8.0 | 0.38 | |
| CRAB C5 | 16 | 64 | 8.0 | 16 | 0.75 | |
| PapMA-3 + Meropenem | CRAB C1 | 16 | 128 | 4.0 | 32 | 0.50 |
| CRAB C2 | 16 | 64 | 8.0 | 32 | 1.00 | |
| CRAB C3 | 16 | 64 | 8.0 | 16 | 0.75 | |
| CRAB C4 | 32 | 128 | 8.0 | 32 | 0.50 | |
| CRAB C5 | 16 | 64 | 8.0 | 16 | 0.75 | |
| PapMA-3 + Rifampin | CRAB C1 | 16 | 128 | 4.0 | 8.0 | 0.31 |
| CRAB C2 | 16 | 64 | 2.0 | 8.0 | 0.25 | |
| CRAB C3 | 16 | 128 | 4.0 | 16 | 0.38 | |
| CRAB C4 | 32 | 128 | 2.0(8.0) | 8.0(4.0) | 0.13(0.27) | |
| CRAB C5 | 16 | 256 | 4.0 | 64 | 0.50 | |
| PapMA-3 + Erythromycin | CRAB C1 | 16 | >512 | 4.0 | 128 | 0.38 |
| CRAB C2 | 16 | 512 | 8.0 | 128 | 0.75 | |
| CRAB C3 | 16 | 512 | 8.0 | 64 | 0.63 | |
| CRAB C4 | 32 | >512 | 4.0(8.0) | 64(16) | 0.19(0.27) | |
| CRAB C5 | 16 | >512 | 4.0 | 256 | 0.50 | |
| PapMA-3 + Vancomycin | CRAB C1 | 16 | 256 | 2.0(4.0) | 32(16) | 0.25(0.31) |
| CRAB C2 | 16 | 256 | 2.0 | 32 | 0.25 | |
| CRAB C3 | 16 | 256 | 8.0 | 4.0 | 0.52 | |
| CRAB C4 | 32 | 512 | 4.0(8.0) | 64(16) | 0.25(0.28) | |
| CRAB C5 | 16 | 256 | 4.0 | 64 | 0.50 | |
| PapMA-3 + Linezolid | CRAB C1 | 16 | 256 | 4.0 | 64 | 0.50 |
| CRAB C2 | 16 | 256 | 4.0 | 64 | 0.50 | |
| CRAB C3 | 16 | 256 | 8.0 | 128 | 1.00 | |
| CRAB C4 | 32 | 512 | 8.0 | 64 | 0.38 | |
| CRAB C5 | 16 | 512 | 4.0 | 128 | 0.50 | |
| Microorganisms | Minimum Inhibitory Concentration (μg·mL−1) | |||||||
|---|---|---|---|---|---|---|---|---|
| PapMA-3 | Melittin | Imipenem | PapMA-3 + Imipenem | |||||
| MH Media | + Serum (50%) | MH Media | + Serum (50%) | MH Media | + Serum (50%) | MH Media | + Serum (50%) | |
| E. coli | 16 | 64 | 8 | 256 | ||||
| A. baumannii | 16 | 64 | 16 | 128 | ||||
| CRAB C1 | 16 | 64 | 16 | 256 | 64 | 64 | 4 + 4 | 16 + 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, J.; Jang, A.; Yoon, Y.K.; Kim, Y. Development of Novel Peptides for the Antimicrobial Combination Therapy against Carbapenem-Resistant Acinetobacter baumannii Infection. Pharmaceutics 2021, 13, 1800. https://doi.org/10.3390/pharmaceutics13111800
Choi J, Jang A, Yoon YK, Kim Y. Development of Novel Peptides for the Antimicrobial Combination Therapy against Carbapenem-Resistant Acinetobacter baumannii Infection. Pharmaceutics. 2021; 13(11):1800. https://doi.org/10.3390/pharmaceutics13111800
Chicago/Turabian StyleChoi, Joonhyeok, Ahjin Jang, Young Kyung Yoon, and Yangmee Kim. 2021. "Development of Novel Peptides for the Antimicrobial Combination Therapy against Carbapenem-Resistant Acinetobacter baumannii Infection" Pharmaceutics 13, no. 11: 1800. https://doi.org/10.3390/pharmaceutics13111800