Nanoencapsulation as a General Solution for Lyophilization of Labile Substrates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression of tES
2.2. Purification of tES and Its Subunits
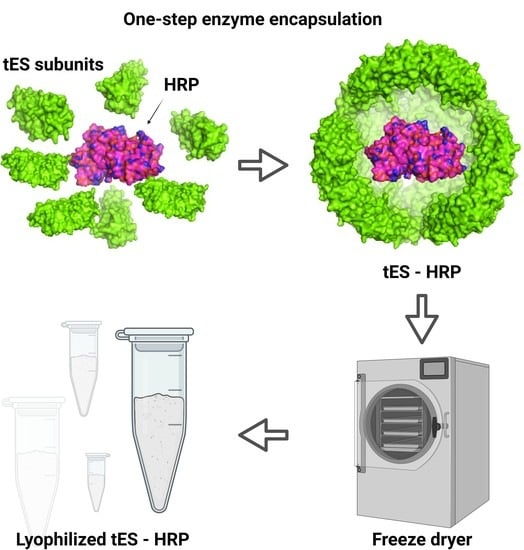
2.3. Nanoencapsulation of HRP within tES
2.4. Size Determination
2.5. CD Spectroscopy
2.6. Lyophilization Experiments
2.7. Thermogravimetric Analysis
2.8. Cytotoxicity Effects
3. Results
3.1. Expression and Purification of tES
3.2. Lyophilization and Stability of Freeze-Dried Proteins
3.3. Nanoencapsulation Studies
3.4. Nanoencapsulation Stabilizes HRP in Dry Form
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dimitrov, D.S. Therapeutic proteins. Ther. Proteins 2012, 899, 1–26. [Google Scholar]
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.-J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS ONE 2017, 12, e0181748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schillberg, S.; Raven, N.; Spiegel, H.; Rasche, S.; Buntru, M. Critical analysis of the commercial potential of plants for the production of recombinant proteins. Front. Plant Sci. 2019, 10, 720. [Google Scholar] [CrossRef]
- Sadeghi, S.; Lee, W.K.; Kong, S.N.; Shetty, A.; Drum, C.L. Oral administration of protein nanoparticles: An emerging route to disease treatment. Pharmacol. Res. 2020, 158, 104685. [Google Scholar] [CrossRef]
- Karki, U.; Fang, H.; Guo, W.; Unnold-Cofre, C.; Xu, J. Cellular engineering of plant cells for improved therapeutic protein production. Plant Cell Rep. 2021, 40, 1087–1099. [Google Scholar] [CrossRef]
- Liu, J.; Nguyen, M.D.; Andya, J.D.; Shire, S.J. Reversible self-association increases the viscosity of a concentrated monoclonal antibody in aqueous solution. J. Pharm. Sci. 2005, 94, 1928–1940. [Google Scholar] [CrossRef]
- Li, L.; Kumar, S.; Buck, P.M.; Burns, C.; Lavoie, J.; Singh, S.K.; Warne, N.W.; Nichols, P.; Luksha, N.; Boardman, D. Concentration dependent viscosity of monoclonal antibody solutions: Explaining experimental behavior in terms of molecular properties. Pharm. Res. 2014, 31, 3161–3178. [Google Scholar] [CrossRef]
- Raut, A.S.; Kalonia, D.S. Pharmaceutical perspective on opalescence and liquid–liquid phase separation in protein solutions. Mol. Pharm. 2016, 13, 1431–1444. [Google Scholar] [CrossRef]
- Sule, S.V.; Cheung, J.K.; Antochshuk, V.; Bhalla, A.S.; Narasimhan, C.; Blaisdell, S.; Shameem, M.; Tessier, P.M. Solution pH that minimizes self-association of three monoclonal antibodies is strongly dependent on ionic strength. Mol. Pharm. 2012, 9, 744–751. [Google Scholar] [CrossRef]
- Chaturvedi, S.K.; Parupudi, A.; Juul-Madsen, K.; Nguyen, A.; Vorup-Jensen, T.; Dragulin-Otto, S.; Zhao, H.; Esfandiary, R.; Schuck, P. Measuring aggregates, self-association, and weak interactions in concentrated therapeutic antibody solutions. In Proceedings of the MAbs Conference, Auckland, New Zealand, 10 May 2020; p. 1810488. [Google Scholar]
- Pham, N.B.; Meng, W.S. Protein aggregation and immunogenicity of biotherapeutics. Int. J. Pharm. 2020, 585, 119523. [Google Scholar] [CrossRef]
- Moussa, E.M.; Panchal, J.P.; Moorthy, B.S.; Blum, J.S.; Joubert, M.K.; Narhi, L.O.; Topp, E.M. Immunogenicity of therapeutic protein aggregates. J. Pharm. Sci. 2016, 105, 417–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W. Instability, stabilization, and formulation of liquid protein pharmaceuticals. Int. J. Pharm. 1999, 185, 129–188. [Google Scholar] [CrossRef]
- Filipe, V.; Hawe, A.; Carpenter, J.F.; Jiskoot, W. Analytical approaches to assess the degradation of therapeutic proteins. TrAC Trends Anal. Chem. 2013, 49, 118–125. [Google Scholar] [CrossRef]
- Tang, X.C.; Pikal, M.J. Design of freeze-drying processes for pharmaceuticals: Practical advice. Pharm. Res. 2004, 21, 191–200. [Google Scholar] [CrossRef]
- Lim, J.Y.; Kim, N.A.; Lim, D.G.; Kim, K.H.; Choi, D.H.; Jeong, S.H. Process cycle development of freeze drying for therapeutic proteins with stability evaluation. J. Pharm. Investig. 2016, 46, 519–536. [Google Scholar] [CrossRef]
- Pieters, S.; de Beer, T.; Kasper, J.C.; Boulpaep, D.; Waszkiewicz, O.; Goodarzi, M.; Tistaert, C.; Friess, W.; Remon, J.-P.; Vervaet, C. Near-infrared spectroscopy for in-line monitoring of protein unfolding and its interactions with lyoprotectants during freeze-drying. Anal. Chem. 2012, 84, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Wang, W. Lyophilization and development of solid protein pharmaceuticals. Int. J. Pharm. 2000, 203, 1–60. [Google Scholar] [CrossRef]
- Faghihi, H.; Khalili, F.; Amini, M.; Vatanara, A. The effect of freeze-dried antibody concentrations on its stability in the presence of trehalose and hydroxypropyl-β-cyclodextrin: A box–behnken statistical design. Pharm. Dev. Technol. 2017, 22, 724–732. [Google Scholar] [CrossRef]
- Kadoya, S.; Fujii, K.; Izutsu, K.-I.; Yonemochi, E.; Terada, K.; Yomota, C.; Kawanishi, T. Freeze-drying of proteins with glass-forming oligosaccharide-derived sugar alcohols. Int. J. Pharm. 2010, 389, 107–113. [Google Scholar] [CrossRef]
- Chang, L.L.; Shepherd, D.; Sun, J.; Ouellette, D.; Grant, K.L.; Tang, X.C.; Pikal, M.J. Mechanism of protein stabilization by sugars during freeze-drying and storage: Native structure preservation, specific interaction, and/or immobilization in a glassy matrix? J. Pharm. Sci. 2005, 94, 1427–1444. [Google Scholar] [CrossRef]
- Mensink, M.A.; Frijlink, H.W.; van der Voort Maarschalk, K.; Hinrichs, W.L. How sugars protect proteins in the solid state and during drying (review): Mechanisms of stabilization in relation to stress conditions. Eur. J. Pharm. Biopharm. 2017, 114, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Kamerzell, T.J.; Esfandiary, R.; Joshi, S.B.; Middaugh, C.R.; Volkin, D.B. Protein–excipient interactions: Mechanisms and biophysical characterization applied to protein formulation development. Adv. Drug Deliv. Rev. 2011, 63, 1118–1159. [Google Scholar] [CrossRef] [PubMed]
- Hudson, K.L.; Bartlett, G.J.; Diehl, R.C.; Agirre, J.; Gallagher, T.; Kiessling, L.L.; Woolfson, D.N. Carbohydrate–aromatic interactions in proteins. J. Am. Chem. Soc. 2015, 137, 15152–15160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Patapoff, T.W.; Overcashier, D.; Hsu, C.; Nguyen, T.H.; Borchardt, R.T. Effects of reducing sugars on the chemical stability of human relaxin in the lyophilized state. J. Pharm. Sci. 1996, 85, 873–877. [Google Scholar] [CrossRef]
- Xu, H.; Templeton, A.C.; A Reed, R. Quantification of 5-HMF and dextrose in commercial aqueous dextrose solutions. J. Pharm. Biomed. Anal. 2003, 32, 451–459. [Google Scholar] [CrossRef]
- Allen, T.M. Drug Delivery Systems: Entering the mainstream. Science 2004, 303, 1818–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, G.; Mitragotri, S.; Tong, S. Multifunctional nanoparticles for drug delivery and molecular imaging. Annu. Rev. Biomed. Eng. 2013, 15, 253–282. [Google Scholar] [CrossRef]
- Hong, S.; Choi, D.W.; Kim, H.N.; Park, C.G.; Lee, W.; Park, H.H. Protein-based nanoparticles as drug delivery systems. Pharmaceutics 2020, 12, 604. [Google Scholar] [CrossRef]
- Franzé, S.; Selmin, F.; Samaritani, E.; Minghetti, P.; Cilurzo, F. Lyophilization of liposomal formulations: Still necessary, still challenging. Pharmaceutics 2018, 10, 139. [Google Scholar] [CrossRef] [Green Version]
- Picco, A.S.; Mondo, G.B.; Ferreira, L.F.; de Souza, E.E.; Peroni, L.A.; Cardoso, M.B. Protein corona meets freeze-drying: Overcoming the challenges of colloidal stability, toxicity, and opsonin adsorption. Nanoscale 2021, 13, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, S.; Masurkar, N.D.; Girish, V.M.; Desai, M.; Chakraborty, G.; Chan, J.M.; Drum, C.L. Thermostable exoshells fold and stabilize recombinant proteins. Nat. Commun. 2017, 8, 1442. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Voss, N.R.; Gerstein, M. 3V: Cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010, 38, W555–W562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franks, F. Freeze-drying of bioproducts: Putting principles into practice. Eur. J. Pharm. BioPharm. 1998, 45, 221–229. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef] [Green Version]
- Fu, B.; Curry, F.-R.E.; Adamson, R.H.; Weinbaum, S. A model for interpreting the tracer labeling of interendothelial clefts. Ann. Biomed. Eng. 1997, 25, 375–397. [Google Scholar] [CrossRef]
- Sana, B.; Johnson, E.; Le Magueres, P.; Criswell, A.; Cascio, D.; Lim, S. The role of nonconserved residues of Archaeoglobus fulgidus ferritin on its unique structure and biophysical properties. J. Biol. Chem. 2013, 288, 32663–32672. [Google Scholar] [CrossRef] [Green Version]
- Giovannelli, L.; Milanesi, A.; Ugazio, E.; Fracchia, L.; Segale, L. Effect of methyl–β–cyclodextrin and trehalose on the freeze–drying and spray–drying of sericin for cosmetic purposes. Pharmaceuticals 2021, 14, 262. [Google Scholar] [CrossRef]
- Carrasquillo, K.G.; Sanchez, C.; Griebenow, K. Relationship between conformational stability and lyophilization-induced structural changes in chymotrypsin. Biotechnol. Appl. Biochem. 2000, 31, 41–53. [Google Scholar] [CrossRef]
- Roy, I.; Gupta, M.N. Freeze-drying of proteins: Some emerging concerns. Biotechnol. Appl. Biochem. 2004, 39, 165–177. [Google Scholar] [CrossRef]
- Sadeghi, S.; Deshpande, S.; Vallerinteavide Mavelli, G.; Aksoyoglu, A.; Bafna, J.; Winterhalter, M.; Kini, R.M.; Lane, D.P.; Drum, C.L. A general approach to protein folding using thermostable exoshells. Nat. Commun. 2021, 12, 5720. [Google Scholar] [CrossRef]
- Krainer, F.W.; Glieder, A. An updated view on horseradish peroxidases: Recombinant production and biotechnological applications. Appl. Microbiol. Biotechnol. 2015, 99, 1611–1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonifert, G.; Folkes, L.; Gmeiner, C.; Dachs, G.; Spadiut, O. Recombinant horseradish peroxidase variants for targeted cancer treatment. Cancer Med. 2016, 5, 1194–1203. [Google Scholar] [CrossRef] [Green Version]
- Sheng, Y.; Wang, K.; Lu, Q.; Ji, P.; Liu, B.; Zhu, J.; Liu, Q.; Sun, Y.; Zhang, J.; Zhou, E.-M. Nanobody-horseradish peroxidase fusion protein as an ultrasensitive probe to detect antibodies against Newcastle disease virus in the immunoassay. J. Nanobiotechnol. 2019, 17, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Bagmi, M.S.; Khan, M.S.; Ismael, M.A.; Al-Senaidy, A.M.; Bacha, A.B.; Husain, F.M.; Alamery, S.F. An efficient methodology for the purification of date palm peroxidase: Stability comparison with horseradish peroxidase (HRP). Saudi J. Biol. Sci. 2019, 26, 301–307. [Google Scholar] [CrossRef]
- Tetter, S.; Hilvert, D. Enzyme encapsulation by a ferritin cage. Angew. Chem. Int. Ed. 2017, 56, 14933–14936. [Google Scholar] [CrossRef]
- Chakraborti, S.; Korpi, A.; Kumar, M.; Stepien, P.; Kostiainen, M.A.; Heddle, J.G. Three-dimensional protein cage array capable of active enzyme capture and artificial chaperone activity. Nano Lett. 2019, 19, 3918–3924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pulsipher, K.W.; Bulos, J.A.; Villegas, J.A.; Saven, J.G.; Dmochowski, I.J. A protein–protein host–guest complex: Thermostable ferritin encapsulating positively supercharged green fluorescent protein. Protein Sci. 2018, 27, 1755–1766. [Google Scholar] [CrossRef] [Green Version]
- Chakraborti, S.; Lin, T.-Y.; Glatt, S.; Heddle, J.G. Enzyme encapsulation by protein cages. RSC Adv. 2020, 10, 13293–13301. [Google Scholar] [CrossRef] [Green Version]
- Duralliu, A.; Matejtschuk, P.; Stickings, P.; Hassall, L.; Tierney, R.; Williams, D.R. The influence of moisture content and temperature on the long-term storage stability of freeze-dried high concentration immunoglobulin G (IgG). Pharmaceutics 2020, 12, 303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Towns, J.K. Moisture content in proteins: Its effects and measurement. J. Chromatogr. A 1995, 705, 115–127. [Google Scholar] [CrossRef]
- May, J.C. Regulatory control of freeze-dried products: Importance and evaluation of residual moisture. In Freeze-Drying/Lyophilization of Pharmaceutical and Biological Products; CRC Press: Boca Raton, FL, USA, 2016; pp. 302–330. [Google Scholar]
- Haeuser, C.; Goldbach, P.; Huwyler, J.; Friess, W.; Allmendinger, A. Imaging techniques to characterize cake appearance of freeze-dried products. J. Pharm. Sci. 2018, 107, 2810–2822. [Google Scholar] [CrossRef] [PubMed]
- Tejedor, M.B.; Fransson, J.; Millqvist-Fureby, A. Freeze-dried cake structural and physical heterogeneity in relation to freeze-drying cycle parameters. Int. J. Pharm. 2020, 590, 119891. [Google Scholar] [CrossRef]
- Patel, S.M.; Nail, S.L.; Pikal, M.J.; Geidobler, R.; Winter, G.; Hawe, A.; Davagnino, J.; Gupta, S.R. Lyophilized drug product cake appearance: What is acceptable? J. Pharm. Sci. 2017, 106, 1706–1721. [Google Scholar] [CrossRef] [PubMed]




| Sample | Td * (°C) Td ** (°C) | Moisture Content (%) |
|---|---|---|
| HRP | 55.5 ± 2.6 307.2 ± 0.1 | 4.6 ± 0.2 |
| tES | 59.7 ± 2.9 328.5 ± 3.0 | 5.5 ± 0.9 |
| tES-HRP | 59.4 ± 3.4 329.5 ± 1.9 | 4.4 ± 0.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vallerinteavide Mavelli, G.; Sadeghi, S.; Vaidya, S.S.; Kong, S.N.; Drum, C.L. Nanoencapsulation as a General Solution for Lyophilization of Labile Substrates. Pharmaceutics 2021, 13, 1790. https://doi.org/10.3390/pharmaceutics13111790
Vallerinteavide Mavelli G, Sadeghi S, Vaidya SS, Kong SN, Drum CL. Nanoencapsulation as a General Solution for Lyophilization of Labile Substrates. Pharmaceutics. 2021; 13(11):1790. https://doi.org/10.3390/pharmaceutics13111790
Chicago/Turabian StyleVallerinteavide Mavelli, Girish, Samira Sadeghi, Siddhesh Sujit Vaidya, Shik Nie Kong, and Chester Lee Drum. 2021. "Nanoencapsulation as a General Solution for Lyophilization of Labile Substrates" Pharmaceutics 13, no. 11: 1790. https://doi.org/10.3390/pharmaceutics13111790