Identification of Effective Anticancer G-Quadruplex-Targeting Chemotypes through the Exploration of a High Diversity Library of Natural Compounds
Abstract
:1. Introduction
2. Results and Discussion
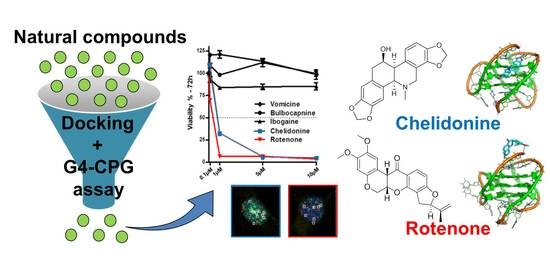
2.1. Docking-Based Virtual Screening of the In-House Library of Natural Compounds
2.2. Experimental Screening of the Library of 28 Natural Compounds by the G4-CPG Assay
2.3. Circular Dichroism Studies
2.4. Fluorescence Spectroscopy Studies
2.5. Molecular Dynamics Simulations
2.6. Evaluation of Biological Activity of the Identified G4 Ligands
3. Conclusions
4. Materials and Methods
4.1. Chemistry
4.2. Molecular Docking
4.3. G4-CPG Assay
4.4. Circular Dichroism
4.5. Fluorescence Spectroscopy
4.6. Molecular Dynamics Simulations
4.7. Biological Assays
4.7.1. Cells and Culture Conditions
4.7.2. Viability Assay (Crystal Violet)
4.7.3. Immunofluorescence (IF) and Fluorescence in situ Hybridization (FISH) Assays
4.7.4. Clonogenic Assay
4.7.5. Western Blot
4.7.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shan, C.; Tan, J.H.; Ou, T.M.; Huang, Z.S. Natural products and their derivatives as G-quadruplex binding ligands. Sci. China Chem. 2013, 56, 1351–1363. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The structure and function of DNA G-quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Hu, S.; Gu, Y.; Yan, Y.; Stovall, D.B.; Li, D.; Sui, G. Human MYC G-quadruplex: From discovery to a cancer therapeutic target. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188410. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-quadruplex, friend or foe: The role of the G-quartet in anticancer strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef]
- Ohnmacht, S.A.; Marchetti, C.; Gunaratnam, M.; Besser, R.J.; Haider, S.M.; Di Vita, G.; Lowe, H.L.; Mellinas-Gomez, M.; Diocou, S.; Robson, M.; et al. A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci. Rep. 2015, 5, 11385. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Dos Santos, N.; Silvester, J.; Wei, V.; Garcia, J.; et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Napolitano, E.; Riccardi, C.; Musumeci, D.; Montesarchio, D. Disentangling the structure–activity relationships of naphthalene diimides as anticancer G-quadruplex-targeting drugs. J. Med. Chem. 2021, 64, 3578–3603. [Google Scholar] [CrossRef]
- Amato, J.; Pagano, A.; Capasso, D.; Di Gaetano, S.; Giustiniano, M.; Novellino, E.; Randazzo, A.; Pagano, B. Targeting the BCL2 gene promoter G-quadruplex with a new class of furopyridazinone-based molecules. ChemMedChem 2018, 13, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Guida, S.; Bonmassar, L.; Aquino, A.; Bonmassar, E.; Ravagnan, G.; Montesarchio, D.; Roviello, G.N.; Musumeci, D.; Fuggetta, M.P. Antitumour activity of resveratrol on human melanoma cells: A possible mechanism related to its interaction with malignant cell telomerase. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2843–2851. [Google Scholar] [CrossRef]
- Platella, C.; Raucci, U.; Rega, N.; D’Atri, S.; Levati, L.; Roviello, G.N.; Fuggetta, M.P.; Musumeci, D.; Montesarchio, D. Shedding light on the interaction of polydatin and resveratrol with G-quadruplex and duplex DNA: A biophysical, computational and biological approach. Int. J. Biol. Macromol. 2020, 151, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Mazzini, S.; Napolitano, E.; Mattio, L.M.; Beretta, G.L.; Zaffaroni, N.; Pinto, A.; Montesarchio, D.; Dallavalle, S. Plant-derived stilbenoids as DNA-binding agents: From monomers to dimers. Chem. Eur. J. 2021, 27, 8832–8845. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, S.; Abbasi, B.H.; Uzair, B.; Abbasi, R. Natural products as reservoirs of novel therapeutic agents. EXCLI J. 2018, 17, 420–451. [Google Scholar] [CrossRef]
- Cappiello, F.; Loffredo, M.R.; Del Plato, C.; Cammarone, S.; Casciaro, B.; Quaglio, D.; Mangoni, M.L.; Botta, B.; Ghirga, F. The revaluation of plant-derived terpenes to fight antibiotic-resistant infections. Antibiotics 2020, 9, 325. [Google Scholar] [CrossRef]
- Casciaro, B.; Mangiardi, L.; Cappiello, F.; Romeo, I.; Loffredo, M.R.; Iazzetti, A.; Calcaterra, A.; Goggiamani, A.; Ghirga, F.; Mangoni, M.L.; et al. Naturally-occurring alkaloids of plant origin as potential antimicrobials against antibiotic-resistant infections. Molecules 2020, 25, 3619. [Google Scholar] [CrossRef] [PubMed]
- Quaglio, D.; Corradi, S.; Erazo, S.; Vergine, V.; Berardozzi, S.; Sciubba, F.; Cappiello, F.; Crestoni, M.E.; Ascenzioni, F.; Imperi, F.; et al. Structural elucidation and antimicrobial characterization of novel diterpenoids from Fabiana densa var. ramulosa. ACS Med. Chem. Lett. 2020, 11, 760–765. [Google Scholar] [CrossRef]
- Chen, Y.; De Bruyn Kops, C.; Kirchmair, J. Data resources for the computer-guided discovery of bioactive natural products. J. Chem. Inf. Model. 2017, 57, 2099–2111. [Google Scholar] [CrossRef]
- Romano, J.D.; Tatonetti, N.P. Informatics and computational methods in natural product drug discovery: A review and perspectives. Front. Genet. 2019, 10, 368. [Google Scholar] [CrossRef] [Green Version]
- Linington, R.G.; Kubanek, J.; Luesch, H. New methods for isolation and structure determination of natural products. Nat. Prod. Rep. 2019, 36, 942–943. [Google Scholar] [CrossRef]
- Feng, W.; Li, M.; Hao, Z.; Zhang, J. Analytical methods of isolation and identification. In Phytochemicals in Human Health; IntechOpen: London, UK, 2019. [Google Scholar]
- Wilson, B.A.P.; Thornburg, C.C.; Henrich, C.J.; Grkovic, T.; O’Keefe, B.R. Creating and screening natural product libraries. Nat. Prod. Rep. 2020, 37, 893–918. [Google Scholar] [CrossRef] [PubMed]
- Ghirga, F.; Quaglio, D.; Mori, M.; Cammarone, S.; Iazzetti, A.; Goggiamani, A.; Ingallina, C.; Botta, B.; Calcaterra, A. A unique high-diversity natural product collection as a reservoir of new therapeutic leads. Org. Chem. Front. 2021, 8, 996–1025. [Google Scholar] [CrossRef]
- Quaglio, D.; Mangoni, M.L.; Stefanelli, R.; Corradi, S.; Casciaro, B.; Vergine, V.; Lucantoni, F.; Cavinato, L.; Cammarone, S.; Loffredo, M.R.; et al. Ent-Beyerane diterpenes as a key platform for the development of ArnT-mediated colistin resistance inhibitors. J. Org. Chem. 2020, 85, 10891–10901. [Google Scholar] [CrossRef]
- Ghirga, F.; Stefanelli, R.; Cavinato, L.; Lo Sciuto, A.; Corradi, S.; Quaglio, D.; Calcaterra, A.; Casciaro, B.; Loffredo, M.R.; Cappiello, F.; et al. A novel colistin adjuvant identified by virtual screening for ArnT inhibitors. J. Antimicrob. Chemother. 2020, 75, 2564–2572. [Google Scholar] [CrossRef] [PubMed]
- Quaglio, D.; Zhdanovskaya, N.; Tobajas, G.; Cuartas, V.; Balducci, S.; Christodoulou, M.S.; Fabrizi, G.; Gargantilla, M.; Priego, E.M.; Carmona Pestania, Á.; et al. Chalcones and chalcone-mimetic derivatives as notch inhibitors in a model of T-cell acute lymphoblastic leukemia. ACS Med. Chem. Lett. 2019, 10, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Lospinoso Severini, L.; Quaglio, D.; Basili, I.; Ghirga, F.; Bufalieri, F.; Caimano, M.; Balducci, S.; Moretti, M.; Romeo, I.; Loricchio, E.; et al. A Smo/Gli multitarget hedgehog oathway inhibitor impairs tumor growth. Cancers 2019, 11, 1518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Infante, P.; Mori, M.; Alfonsi, R.; Ghirga, F.; Aiello, F.; Toscano, S.; Ingallina, C.; Siler, M.; Cucchi, D.; Po, A.; et al. Gli1/DNA interaction is a druggable target for Hedgehog-dependent tumors. EMBO J. 2015, 34, 200–217. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.; Jones, R.A.; Yang, D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry 2005, 44, 2048–2058. [Google Scholar] [CrossRef]
- Dai, J.; Carver, M.; Punchihewa, C.; Jones, R.A.; Yang, D. Structure of the hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: Insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007, 35, 4927–4940. [Google Scholar] [CrossRef] [Green Version]
- Neidle, S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016, 59, 5987–6011. [Google Scholar] [CrossRef]
- He, X.; Zhang, Z.; Zhang, Q.; Yuan, G. Selective recognition of G-quadruplex in the vascular endothelial growth factor gene with small-molecule natural products by electrospray ionization (ESI) mass spectrometry and circular dichroism (CD) spectrometry. Can. J. Chem. 2012, 90, 55–59. [Google Scholar] [CrossRef]
- Noureini, S.K.; Esmaeili, H.; Abachi, F.; Khiali, S.; Islam, B.; Kuta, M.; Saboury, A.A.; Hoffmann, M.; Sponer, J.; Parkinson, G.; et al. Selectivity of major isoquinoline alkaloids from Chelidonium majus towards telomeric G-quadruplex: A study using a transition-FRET (t-FRET) assay. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2020–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirota, V.; Platella, C.; Musumeci, D.; Benassi, A.; Amato, J.; Pagano, B.; Colombo, G.; Freccero, M.; Doria, F.; Montesarchio, D. On the binding of naphthalene diimides to a human telomeric G-quadruplex multimer model. Int. J. Biol. Macromol. 2020, 166, 1320–1334. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Pirota, V.; Musumeci, D.; Rizzi, F.; Iachettini, S.; Zizza, P.; Biroccio, A.; Freccero, M.; Montesarchio, D.; Doria, F. Trifunctionalized naphthalene diimides and dimeric analogues as G-quadruplex-targeting anticancer agents selected by affinity chromatography. Int. J. Mol. Sci. 2020, 21, 1964. [Google Scholar] [CrossRef] [Green Version]
- Amato, J.; Platella, C.; Iachettini, S.; Zizza, P.; Musumeci, D.; Cosconati, S.; Pagano, A.; Novellino, E.; Biroccio, A.; Randazzo, A.; et al. Tailoring a lead-like compound targeting multiple G-quadruplex structures. Eur. J. Med. Chem. 2019, 163, 295–306. [Google Scholar] [CrossRef]
- Platella, C.; Musumeci, D.; Arciello, A.; Doria, F.; Freccero, M.; Randazzo, A.; Amato, J.; Pagano, B.; Montesarchio, D. Controlled Pore Glass-based oligonucleotide affinity support: Towards High Throughput Screening methods for the identification of conformation-selective G-quadruplex ligands. Anal. Chim. Acta 2018, 1030, 133–141. [Google Scholar] [CrossRef]
- Musumeci, D.; Amato, J.; Zizza, P.; Platella, C.; Cosconati, S.; Cingolani, C.; Biroccio, A.; Novellino, E.; Randazzo, A.; Giancola, C.; et al. Tandem application of ligand-based virtual screening and G4-OAS assay to identify novel G-quadruplex-targeting chemotypes. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1341–1352. [Google Scholar] [CrossRef]
- Platella, C.; Trajkovski, M.; Doria, F.; Freccero, M.; Montesarchio, D.; Plavec, J. On the interaction of an anticancer trisubstituted naphthalene diimide with G-quadruplexes of different topologies: A structural insight. Nucleic Acids Res. 2020, 48, 12380–12393. [Google Scholar] [CrossRef]
- Mori, M.; Tottone, L.; Quaglio, D.; Zhdanovskaya, N.; Ingallina, C.; Fusto, M.; Ghirga, F.; Peruzzi, G.; Crestoni, M.E.; Simeoni, F.; et al. Identification of a novel chalcone derivative that inhibits Notch signaling in T-cell acute lymphoblastic leukemia. Sci. Rep. 2017, 7, 2213. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug. Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [Green Version]
- del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-quadruplex secondary structure obtained from circular dichroism spectroscopy. Angew. Chem. Int. Ed. 2018, 57, 7171–7175. [Google Scholar] [CrossRef]
- Kypr, J.; Kejnovská, I.; Renčiuk, D.; Vorlíčková, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [Green Version]
- Giancola, C.; Pagano, B. Energetics of ligand binding to G-quadruplexes. Top. Curr. Chem. 2013, 330, 211–242. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Rodriguez, R.; Müller, S.; Yeoman, J.A.; Trentesaux, C.; Riou, J.F.; Balasubramanian, S. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008, 130, 15758–15759. [Google Scholar] [CrossRef] [Green Version]
- Francisco, S.; Francisco, S. γ-H2AX—A novel biomarker for DNA double-strand breaks. In Vivo 2008, 22, 305–309. [Google Scholar]
- Rizzo, A.; Iachettini, S.; Zizza, P.; Cingolani, C.; Porru, M.; Artuso, S.; Stevens, M.; Hummersone, M.; Biroccio, A.; Salvati, E.; et al. Identification of novel RHPS4-derivative ligands with improved toxicological profiles and telomere-targeting activities. J. Exp. Clin. Cancer Res. 2014, 33, 81. [Google Scholar] [CrossRef] [PubMed]
- Denchi, E.L.; De Lange, T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007, 448, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Van Steensel, B.; Smogorzewska, A.; De Lange, T. TRF2 protects human telomeres from end-to-end fusions. Cell 1998, 92, 401–413. [Google Scholar] [CrossRef] [Green Version]
- Qian, L.; Chen, K.; Wang, C.; Chen, Z.; Meng, Z.; Wang, P. Targeting NRAS-mutant cancers with the selective STK19 kinase inhibitor chelidonine. Clin. Cancer Res. 2020, 26, 3408–3419. [Google Scholar] [CrossRef] [Green Version]
- Xiao, W.; Liu, Y.; Dai, M.; Li, Y.; Peng, R.; Yu, S.; Liu, H. Rotenone restrains colon cancer cell viability, motility and epithelial-mesenchymal transition and tumorigenesis in nude mice via the PI3K/AKT pathway. Int. J. Mol. Med. 2020, 46, 700–708. [Google Scholar] [CrossRef]
- Guimarães, H.A.; Braz-Filho, R.; Vieira, I.J.C. 1H and 13C-NMR data of the simplest plumeran indole alkaloids isolated from Aspidosperma species. Molecules 2012, 17, 3025–3043. [Google Scholar] [CrossRef] [Green Version]
- Alper, K.R. Ibogaine: A review. Alkaloids. Chem. Biol. 2001, 56, 1–38. [Google Scholar]
- Niitsu, A.; Harada, M.; Yamagaki, T.; Tachibana, K. Conformations of 3-carboxylic esters essential for neurotoxicity in veratrum alkaloids are loosely restricted and fluctuate. Bioorg. Med. Chem. 2008, 16, 3025–3031. [Google Scholar] [CrossRef] [PubMed]
- Monache, F.D.; Mc Quhae, M.M.; Ferrari, F.; Marini-Bettolo, G.B. Ferruginin A and B and ferruanthrone, new triprenylated anthranoids from Vismia baccifera var. ferruginea. Tetrahedron 1979, 35, 2143–2149. [Google Scholar] [CrossRef]
- Monache, F.D.; Torres, F.F.; Marini-Bettolo, G.B.; De Lima, R.A. Chemistry of Vismia Genus. Note V: γ-Hydroxy-and γ, γ’-Dihydroxy-Ferruginin A. J. Nat. Prod. 1980, 43, 487–494. [Google Scholar] [CrossRef]
- Nicoletti, M.; Marini-Bettolo, G.B.; Delle Monache, F.; Delle Monache, G. Keto-enolic tautomerism and spectral data of prenylated anthranoids from Vismia genus. Tetrahedron 1982, 38, 3679–3686. [Google Scholar] [CrossRef]
- Manitto, P.; Monti, D.; Speranza, G.; Mulinacci, N.; Vincieri, F.F.; Griffini, A.; Pifferi, G. Conformational studies of natural products. Part 4. Conformation and absolute configuration of cascarosides A, B, C, D. J. Chem. Soc. Perkin Trans. 1 1993, 1577–1580. [Google Scholar] [CrossRef]
- Delle Monache, A.; Ferrari, F.; Marini Bettolo, G.B. Vismiones from Vismia baccifera var. dealdata (H.B.K.): Chemistry and X-ray structure determination. Gazz. Chim. Ital. 1979, 109, 301–310. [Google Scholar]
- Russell, D.A.; Bridges, H.R.; Serreli, R.; Kidd, S.L.; Mateu, N.; Osberger, T.J.; Sore, H.F.; Hirst, J.; Spring, D.R. Hydroxylated rotenoids selectively inhibit the proliferation of prostate cancer cells. J. Nat. Prod. 2020, 83, 1829–1845. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, J.G.; Olivares, E.M.; Monache, F.D. Citrans and cyclols from Clusia multiflora. Phytochemistry 1995, 38, 485–489. [Google Scholar] [CrossRef]
- Carrillo-Cocom, L.M.; González, B.B.V.; Santillan, R.; Soto-Castro, D.; Ocampo, P.M.S.; Zepeda, A.; Tafur, J.C. Synthesis of diosgenin prodrugs: Anti-inflammatory and antiproliferative activity evaluation. J. Chem. Sci. 2020, 132, 104. [Google Scholar] [CrossRef]
- Zughdani, M.; Yusufoğlu, H.S.; Ekiz, G.; Linden, A.; Çalış, İ. Ecdysteroids from the underground parts of Rhaponticum acaule (L.) DC. Phytochemistry 2020, 180, 112530. [Google Scholar] [CrossRef]
- Hano, Y.; Suzuki, S.; Kohno, H.; Nomura, T. Absolute configuration of kuwanon L, a natural Diels-Alder type adduct from the Morus root bark. Heterocycles 1988, 27, 75–81. [Google Scholar] [CrossRef]
- Bunyapaiboonsri, T.; Yoiprommarat, S.; Suntivich, R.; Preedanon, S.; Komwijit, S.; Teerawatananond, T.; Sakayaroj, J. A cyclic lipodepsipeptide, a spirolactone, and a chromanone from the marine fungus Verruculina enalia (Kohlm.) Kohlm. & Volkm.-Kohlm. BCC 22226. Tetrahedron 2020, 76, 131497. [Google Scholar] [CrossRef]
- Nahrstedt, A.; Sattar, E.A.; El-Zalabani, S.M. Amygdalin acyl derivatives, cyanogenic glycosides from the seeds of Merremia dissecta. Phytochemistry 1990, 29, 3934–3936. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Delaglio, F.; Tjandra, N.; Zhurkin, V.B.; Bax, A. Overall structure and sugar dynamics of a DNA dodecamer from homo and heteronuclear dipolar couplings and 31P chemical shift anisotropy. J. Biomol. NMR 2003, 26, 297–315. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Stahl, M.; Mauser, H. Database clustering with a combination of fingerprint and maximum common substructure methods. J. Chem. Inf. Model. 2005, 45, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Galindo-Murillo, R.; Robertson, J.C.; Zgarbová, M.; Šponer, J.; Otyepka, M.; Jurečka, P.; Cheatham, T.E. Assessing the current state of Amber force field modifications for DNA. J. Chem. Theory Comput. 2016, 12, 4114–4127. [Google Scholar] [CrossRef]
- Zgarbová, M.; Šponer, J.; Otyepka, M.; Cheatham, T.E.; Galindo-Murillo, R.; Jurečka, P. Refinement of the sugar-phosphate backbone torsion beta for AMBER force fields improves the description of Z- and B-DNA. J. Chem. Theory Comput. 2015, 11, 5723–5736. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general Amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Mély, Y.; Kuchlyan, J.; Martinez-Fernandez, L.; Mori, M.; Gavvala, K.; Ciaco, S.; Boudier, C.; Richert, L.; Didier, P.; Tor, Y.; et al. What makes thienoguanosine an outstanding fluorescent DNA probe? J. Am. Chem. Soc. 2020, 142, 16999–17014. [Google Scholar] [CrossRef]
- Ballone, A.; Picarazzi, F.; Prosser, C.; Davis, J.; Ottmann, C.; Mori, M. Experimental and computational druggability exploration of the 14-3-3ζ/SOS1pS1161PPI interface. J. Chem. Inf. Model. 2020, 60, 6555–6565. [Google Scholar] [CrossRef]
- Boccuto, A.; Dragoni, F.; Picarazzi, F.; Lai, A.; Della Ventura, C.; Veo, C.; Giammarino, F.; Saladini, F.; Zehender, G.; Zazzi, M.; et al. Sofosbuvir selects for drug-resistant amino acid variants in the zika virus RNA-dependent RNA-polymerase complex in vitro. Int. J. Mol. Sci. 2021, 22, 2670. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3097. [Google Scholar] [CrossRef]
- Chen, W.; Hahn, W.C. SV40 early region oncoproteins and human cell transformation. Histol. Histopathol. 2003, 18, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Bartocci, C.; Ouzounov, I.; Diedrich, J.K.; Yates, J.R.; Denchi, E.L. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 2013, 494, 502–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Percentage of Bound Ligand * | Selectivity Index | |||||
|---|---|---|---|---|---|---|
| Nude CPG | CPG-tel26 | CPG-c-myc | CPG-ds27 | CPG-tel26 vs. CPG-ds27 | CPG-c-myc vs. CPG-ds27 | |
| 20-OH-Ecdysone | ● | ● | ● | ● | 0 | >2 |
| Aloin | ● | ● | ● | ● | >2 | >2 |
| Aspidospermine | ● | ● | ● | ● | 2.5 | 3.0 |
| Bulbocapnine | ● | ● | ● | ● | 1.1 | 2.3 |
| Chelidonine | ● | ● | ● | ● | 1.6 | 0.5 |
| Emetine | ● | ● | ● | ● | 0.3 | 0.7 |
| Ferruanthrone | ● | ● | ● | ● | 1.0 | 1.6 |
| Ferruginin A | ● | ● | ● | ● | 0.9 | 1.5 |
| Ferruginin B | ● | ● | ● | ● | 0 | 0 |
| γ,γ’-OH-Ferruginin A | ● | ● | ● | ● | 0 | 0 |
| Hydrastine | ● | ● | ● | ● | 1.2 | 1.1 |
| Ibogaine | ● | ● | ● | ● | 4.0 | 0 |
| Jervine | ● | ● | ● | ● | 1.1 | 0.9 |
| Kuwanon G | ● | ● | ● | ● | 2.7 | 1.5 |
| Narceine | ● | ● | ● | ● | 1.3 | 0.3 |
| Rotenolone | ● | ● | ● | ● | 0 | 0 |
| Rotenone | ● | ● | ● | ● | 1.4 | 0.6 |
| Veratrine | ● | ● | ● | ● | >2 | 0 |
| Vindoline | ● | ● | ● | ● | 1.0 | 0.1 |
| Vismione B | ● | ● | ● | ● | 0 | 0 |
| Vomicine | ● | ● | ● | ● | 3.0 | 2.3 |
| Yohimbine | ● | ● | ● | ● | >10 | 0 |
| tel26 * | c-myc ** | ds27 * | ||||
|---|---|---|---|---|---|---|
| Tm (°C) (±1) | ΔTm (°C) | Tm (°C) (±1) | ΔTm (°C) | Tm (°C) (±1) | ΔTm (°C) | |
| No ligand | 40 | 45 | 75 | |||
| Bulbocapnine | 41 | +1 | 54 | +9 | 74 | −1 |
| Chelidonine | 44 | +4 | 58 | +13 | 74 | −1 |
| Ibogaine | 42 | +2 | 50 | +5 | 75 | 0 |
| Rotenone | 40 | 0 | 49 | +4 | 75 | 0 |
| Vomicine | 40 | 0 | 50 | +5 | 72 | −3 |
| Kb (M−1) | |||
|---|---|---|---|
| tel26 | c-myc | ds27 | |
| Bulbocapnine | 1.0 ( ± 0.4) × 106 | 1.1 ( ± 0.4) × 106 | 1.2 ( ± 0.6) × 106 |
| Ibogaine | 4.1 ( ± 0.8) × 105 | 3.9 ( ± 0.7) × 105 | 3.7 ( ± 0.7) × 105 |
| Compound | tel26 (MM-GBSA) kcal/mol ± SEM | c-myc (MM-GBSA) kcal/mol ± SEM |
|---|---|---|
| Bulbocapnine | −16.85 ± 0.43 | −38.57 ± 0.35 |
| Chelidonine | −17.49 ± 0.31 | −25.07 ± 0.42 |
| Ibogaine | −23.92 ± 0.34 | −21.56 ± 0.71 |
| Rotenone | −4.47 ± 0.73 | −9.47 ± 0.42 |
| Vomicine | −20.52 ± 0.33 | −12.81 ± 0.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Platella, C.; Ghirga, F.; Zizza, P.; Pompili, L.; Marzano, S.; Pagano, B.; Quaglio, D.; Vergine, V.; Cammarone, S.; Botta, B.; et al. Identification of Effective Anticancer G-Quadruplex-Targeting Chemotypes through the Exploration of a High Diversity Library of Natural Compounds. Pharmaceutics 2021, 13, 1611. https://doi.org/10.3390/pharmaceutics13101611
Platella C, Ghirga F, Zizza P, Pompili L, Marzano S, Pagano B, Quaglio D, Vergine V, Cammarone S, Botta B, et al. Identification of Effective Anticancer G-Quadruplex-Targeting Chemotypes through the Exploration of a High Diversity Library of Natural Compounds. Pharmaceutics. 2021; 13(10):1611. https://doi.org/10.3390/pharmaceutics13101611
Chicago/Turabian StylePlatella, Chiara, Francesca Ghirga, Pasquale Zizza, Luca Pompili, Simona Marzano, Bruno Pagano, Deborah Quaglio, Valeria Vergine, Silvia Cammarone, Bruno Botta, and et al. 2021. "Identification of Effective Anticancer G-Quadruplex-Targeting Chemotypes through the Exploration of a High Diversity Library of Natural Compounds" Pharmaceutics 13, no. 10: 1611. https://doi.org/10.3390/pharmaceutics13101611