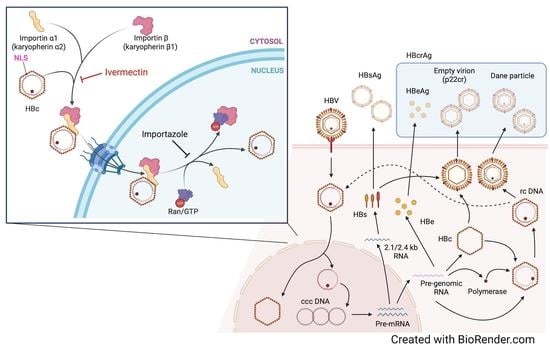
Ivermectin Inhibits HBV Entry into the Nucleus by Suppressing KPNA2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Compounds
2.2. Cell Culture
2.3. Infection of HepG2-hNTCP-C4 Cells with HBV
2.4. Infection of PXB Hepatocytes with HBV
2.5. Cytotoxicity Assay
2.6. HBV DNA Quantification by Quantitative Polymerase Chain Reaction (qPCR)
2.7. cccDNA Quantification by Droplet Digital Polymerase Chain Reaction (ddPCR)
2.8. Measurement of HBsAg and Hepatitis B Core-Related Antigen (HBcrAg) Levels in Culture Supernatants
2.9. PreS1 Binding Assay
2.10. Hepatitis B Core Protein (HBc) Plasmid Transfection
2.11. Small Interfering RNA (siRNA) Transfection
2.12. Reverse Transcription Real-Time Quantitative PCR
2.13. Immunofluorescence Staining
2.14. Western Blotting and Simple Western Assay
2.15. Statistical Analysis
3. Results
3.1. Ivermectin Inhibits HBV Infection in HepG2-hNTCP-C4 Cells
3.2. Anti-HBV Effects of Ivermectin in the Initial HBV Infection in PXB Hepatocytes
3.3. Ivermectin Inhibits HBV Infection via a Novel Mechanism
3.4. Ivermectin Inhibits the Nuclear Transfer of HBc
3.5. Ivermectin Decreases the Nuclear Localization of KPNA2
3.6. Multiple Subtypes of Importin α Are Involved in HBV Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. World Health Organization Hepatitis B-Key Facts. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (accessed on 5 July 2022).
- McMahon, B.J. Epidemiology and Natural History of Hepatitis B. Semin. Liver Dis. 2005, 25, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Block, T.M.; Guo, H.; Guo, J.-T. Molecular Virology of Hepatitis B Virus for Clinicians. Clin. Liver Dis. 2007, 11, 685–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, T.J. Hepatitis B: The virus and disease. Hepatology 2009, 49, 13–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seto, W.K.; Lo, Y.R.; Pawlotsky, J.M.; Yuen, M.F. Chronic hepatitis B virus infection. Lancet 2018, 392, 2313–2324. [Google Scholar] [CrossRef]
- Sarin, S.K.; Kumar, M.; Lau, G.K.; Abbas, Z.; Chan, H.L.; Chen, C.J.; Chen, D.S.; Chen, H.L.; Chen, P.J.; Chien, R.N.; et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: A 2015 update. Hepatol. Int. 2016, 10, 1–98. [Google Scholar] [CrossRef]
- Terrault, N.A.; Bzowej, N.H.; Chang, K.M.; Hwang, J.P.; Jonas, M.M.; Murad, M.H.; American Association for the Study of Liver Diseases. AASLD guidelines for treatment of chronic hepatitis B. Hepatology 2016, 63, 261–283. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017, 67, 370–398. [Google Scholar] [CrossRef] [Green Version]
- Tseng, T.C.; Liu, C.J.; Yang, H.C.; Su, T.H.; Wang, C.C.; Chen, C.L.; Kuo, S.F.; Liu, C.H.; Chen, P.J.; Chen, D.S.; et al. High levels of hepatitis B surface antigen increase risk of hepatocellular carcinoma in patients with low HBV load. Gastroenterology 2012, 142, 1140–1149.e1143. [Google Scholar] [CrossRef] [Green Version]
- Chien, R.N.; Liaw, Y.F. Current trend in antiviral therapy for chronic hepatitis B. Viruses 2022, 14, 434. [Google Scholar] [CrossRef]
- Werle-Lapostolle, B.; Bowden, S.; Locarnini, S.; Wursthorn, K.; Petersen, J.; Lau, G.; Trepo, C.; Marcellin, P.; Goodman, Z.; Delaney, W.E., 4th; et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology 2004, 126, 1750–1758. [Google Scholar] [CrossRef]
- Martinez, M.G.; Testoni, B.; Zoulim, F. Biological basis for functional cure of chronic hepatitis B. J. Viral Hepat. 2019, 26, 786–794. [Google Scholar] [CrossRef]
- Papatheodoridis, G.; Vlachogiannakos, I.; Cholongitas, E.; Wursthorn, K.; Thomadakis, C.; Touloumi, G.; Petersen, J. Discontinuation of oral antivirals in chronic hepatitis B: A systematic review. Hepatology 2016, 63, 1481–1492. [Google Scholar] [CrossRef]
- Ghany, M.G.; Doo, E.C. Antiviral resistance and hepatitis B therapy. Hepatology 2009, 49, 174–184. [Google Scholar] [CrossRef] [Green Version]
- Janssen, H.L.; van Zonneveld, M.; Senturk, H.; Zeuzem, S.; Akarca, U.S.; Cakaloglu, Y.; Simon, C.; So, T.M.; Gerken, G.; de Man, R.A.; et al. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: A randomised trial. Lancet 2005, 365, 123–129. [Google Scholar] [CrossRef]
- Lau, G.K.; Piratvisuth, T.; Luo, K.X.; Marcellin, P.; Thongsawat, S.; Cooksley, G.; Gane, E.; Fried, M.W.; Chow, W.C.; Paik, S.W.; et al. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N. Engl. J. Med. 2005, 352, 2682–2695. [Google Scholar] [CrossRef] [Green Version]
- Marcellin, P.; Lau, G.K.; Bonino, F.; Farci, P.; Hadziyannis, S.; Jin, R.; Lu, Z.M.; Piratvisuth, T.; Germanidis, G.; Yurdaydin, C.; et al. Peginterferon alfa-2a alone, lamivudine alone, and the two in combination in patients with HBeAg-negative chronic hepatitis B. N. Engl. J. Med. 2004, 351, 1206–1217. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.; Hayashi, S.; Tanaka, Y. Drug discovery study aimed at a functional cure for HBV. Viruses 2022, 14, 1393. [Google Scholar] [CrossRef]
- Ko, C.; Bester, R.; Zhou, X.; Xu, Z.; Blossey, C.; Sacherl, J.; Vondran, F.W.R.; Gao, L.; Protzer, U. A new role for capsid assembly modulators to target mature hepatitis B virus capsids and prevent virus infection. Antimicrob. Agents Chemother. 2019, 64, e01440-19. [Google Scholar] [CrossRef]
- Kobayashi, C.; Watanabe, Y.; Oshima, M.; Hirose, T.; Yamasaki, M.; Iwamoto, M.; Iwatsuki, M.; Asami, Y.; Kuramochi, K.; Wakae, K.; et al. Fungal secondary metabolite exophillic acid selectively inhibits the entry of hepatitis B and D viruses. Viruses 2022, 14, 764. [Google Scholar] [CrossRef]
- Seeger, C.; Mason, W.S. Molecular biology of hepatitis B virus infection. Virology 2015, 479, 672–686. [Google Scholar] [CrossRef]
- Hu, J.; Liu, K. Complete and incomplete hepatitis B virus particles: Formation, function, and application. Viruses 2017, 9, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 2012, 1, e00049. [Google Scholar] [CrossRef] [PubMed]
- Tuttleman, J.S.; Pourcel, C.; Summers, J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 1986, 47, 451–460. [Google Scholar] [CrossRef]
- Rall, L.B.; Standring, D.N.; Laub, O.; Rutter, W.J. Transcription of hepatitis B virus by RNA polymerase II. Mol. Cell. Biol. 1983, 3, 1766–1773. [Google Scholar] [PubMed] [Green Version]
- Cautain, B.; Hill, R.; de Pedro, N.; Link, W. Components and regulation of nuclear transport processes. FEBS J. 2015, 282, 445–462. [Google Scholar] [CrossRef]
- Rexach, M.; Blobel, G. Protein import into nuclei: Association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 1995, 83, 683–692. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, Y.; Boag, P.R.; Hime, G.R.; Loveland, K.L. Regulated nucleocytoplasmic transport during gametogenesis. Biochim. Biophys. Acta 2012, 1819, 616–630. [Google Scholar] [CrossRef]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [Green Version]
- Görlich, D.; Kostka, S.; Kraft, R.; Dingwall, C.; Laskey, R.A.; Hartmann, E.; Prehn, S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 1995, 5, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Eckhardt, S.G.; Milich, D.R.; McLachlan, A. Hepatitis B virus core antigen has two nuclear localization sequences in the arginine-rich carboxyl terminus. J. Virol. 1991, 65, 575–582. [Google Scholar] [CrossRef]
- Rabe, B.; Vlachou, A.; Pante, N.; Helenius, A.; Kann, M. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc. Natl. Acad. Sci. USA 2003, 100, 9849–9854. [Google Scholar] [CrossRef] [Green Version]
- Kelley, J.B.; Talley, A.M.; Spencer, A.; Gioeli, D.; Paschal, B.M. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 2010, 11, 63. [Google Scholar] [CrossRef] [Green Version]
- Oka, M.; Yoneda, Y. Importin alpha: Functions as a nuclear transport factor and beyond. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2018, 94, 259–274. [Google Scholar] [CrossRef] [Green Version]
- Crump, A. Ivermectin: Enigmatic multifaceted ‘wonder’ drug continues to surprise and exceed expectations. J. Antibit. 2017, 70, 495–505. [Google Scholar] [CrossRef] [Green Version]
- Campbell, W.C.; Fisher, M.H.; Stapley, E.O.; Alberes-Schonberg, G.; Jacob, T.A. Ivermectin: A potent new antiparasitic agents. Science 1983, 221, 823–828. [Google Scholar] [CrossRef]
- Hibbs, R.E.; Gouaux, E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 2011, 474, 54–60. [Google Scholar] [CrossRef] [Green Version]
- Burkhart, C.N. Ivermectin: An assessment of its pharmacology, microbiology and safety. Vet. Hum. Toxicol. 2000, 42, 30–35. [Google Scholar]
- Henriquez-Camacho, C.; Gotuzzo, E.; Echevarria, J.; White, A.C., Jr.; Terashima, A.; Samalvides, F.; Perez-Molina, J.A.; Plana, M.N. Ivermectin versus albendazole or thiabendazole for Strongyloides stercoralis infection. Cochrane Database Syst. Rev. 2016, 2016, CD007745. [Google Scholar] [CrossRef] [Green Version]
- Rosumeck, S.; Nast, A.; Dressler, C. Ivermectin and permethrin for treating scabies. Cochrane Database Syst. Rev. 2018, 2018, CD012994. [Google Scholar] [CrossRef] [Green Version]
- Abegunde, A.T.; Ahuja, R.M.; Okafor, N.J. Doxycycline plus ivermectin versus ivermectin alone for treatment of patients with onchocerciasis. Cochrane Database Syst. Rev. 2016, 2016, CD011146. [Google Scholar] [CrossRef] [Green Version]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Watashi, K.; Tsukuda, S.; Aly, H.H.; Fukasawa, M.; Fujimoto, A.; Suzuki, R.; Aizaki, H.; Ito, T.; Koiwai, O.; et al. Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochem. Biophys. Res. Commun. 2014, 443, 808–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogura, N.; Watashi, K.; Noguchi, T.; Wakita, T. Formation of covalently closed circular DNA in Hep38.7-Tet cells, a tetracycline inducible hepatitis B virus expression cell line. Biochem. Biophys. Res. Commun. 2014, 452, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Inoue, K.; Hayashi, Y.; Abe, A.; Tsukiyama-Kohara, K.; Nuriya, H.; Aoki, Y.; Kawaguchi, R.; Kubota, K.; Yoshiba, M.; et al. Virological significance of low-level hepatitis B virus infection in patients with hepatitis C virus associated liver disease. J. Med. Virol. 2004, 72, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Hamada-Tsutsumi, S.; Naito, Y.; Sato, S.; Takaoka, A.; Kawashima, K.; Isogawa, M.; Ochiya, T.; Tanaka, Y. The antiviral effects of human microRNA miR-302c-3p against hepatitis B virus infection. Aliment. Pharmacol. Ther. 2019, 49, 1060–1070. [Google Scholar] [CrossRef]
- Zhong, Y.; Hu, S.; Xu, C.; Zhao, Y.; Xu, D.; Zhao, Y.; Zhao, J.; Li, Z.; Zhang, X.; Zhang, H.; et al. A novel method for detection of HBVcccDNA in hepatocytes using rolling circle amplification combined with in situ PCR. BMC Infect. Dis. 2014, 14, 608. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, S.; Isogawa, M.; Kawashima, K.; Ito, K.; Chuaypen, N.; Morine, Y.; Shimada, M.; Higashi-Kuwata, N.; Watanabe, T.; Tangkijvanich, P.; et al. Droplet digital PCR assay provides intrahepatic HBV cccDNA quantification tool for clinical application. Sci. Rep. 2022, 12, 2133. [Google Scholar] [CrossRef]
- Dyavar, S.R.; Ye, Z.; Byrareddy, S.N.; Scarsi, K.K.; Winchester, L.C.; Weinhold, J.A.; Fletcher, C.V.; Podany, A.T. Normalization of cell associated antiretroviral drug concentrations with a novel RPP30 droplet digital PCR assay. Sci. Rep. 2018, 8, 3626. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, M.; Watashi, K.; Kamisuki, S.; Matsunaga, H.; Iwamoto, M.; Kawai, F.; Ohashi, H.; Tsukuda, S.; Shimura, S.; Suzuki, R.; et al. A novel tricyclic polyketide, vanitaracin A, specifically inhibits the entry of hepatitis B and D viruses by targeting sodium taurocholate cotransporting polypeptide. J. Virol. 2015, 89, 11945–11953. [Google Scholar] [CrossRef] [Green Version]
- Kawashima, K.; Isogawa, M.; Hamada-Tsutsumi, S.; Baudi, I.; Saito, S.; Nakajima, A.; Tanaka, Y. Type I interferon signaling prevents hepatitis B virus-specific T cell responses by reducing antigen expression. J. Virol. 2018, 92, e01099-18. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, M.; Tanaka, Y.; Kato, T.; Orito, E.; Ito, K.; Acharya, S.K.; Gish, R.G.; Kramvis, A.; Shimada, T.; Izumi, N.; et al. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology 2006, 44, 915–924. [Google Scholar] [CrossRef]
- Okumura, H.; Nakanishi, A.; Hashita, T.; Iwao, T.; Matsunaga, T. Effect of celecoxib on differentiation of human induced pluripotent stem cells into hepatocytes involves STAT5 activation. Drug Metab. Dispos. 2018, 46, 1519–1527. [Google Scholar] [CrossRef]
- Watashi, K.; Sluder, A.; Daito, T.; Matsunaga, S.; Ryo, A.; Nagamori, S.; Iwamoto, M.; Nakajima, S.; Tsukuda, S.; Borroto-Esoda, K.; et al. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP). Hepatology 2014, 59, 1726–1737. [Google Scholar] [CrossRef] [Green Version]
- Lv, C.; Liu, W.; Wang, B.; Dang, R.; Qiu, L.; Ren, J.; Yan, C.; Yang, Z.; Wang, X. Ivermectin inhibits DNA polymerase UL42 of pseudorabies virus entrance into the nucleus and proliferation of the virus in vitro and vivo. Antiviral. Res. 2018, 159, 55–62. [Google Scholar] [CrossRef]
- Ko, C.; Chakraborty, A.; Chou, W.M.; Hasreiter, J.; Wettengel, J.M.; Stadler, D.; Bester, R.; Asen, T.; Zhang, K.; Wisskirchen, K.; et al. Hepatitis B virus genome recycling and de novo secondary infection events maintain stable cccDNA levels. J. Hepatol. 2018, 69, 1231–1241. [Google Scholar] [CrossRef]
- Levrero, M.; Pollicino, T.; Petersen, J.; Belloni, L.; Raimondo, G.; Dandri, M. Control of cccDNA function in hepatitis B virus infection. J. Hepatol. 2009, 51, 581–592. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Qiu, S.; Lu, M.; Huang, C.; Lv, Y. Nuclear transporter karyopherin subunit alpha 3 levels modulate Porcine circovirus type 2 replication in PK-15 cells. Virology 2020, 548, 31–38. [Google Scholar] [CrossRef]
- Hudjetz, B.; Gabriel, G. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-alpha1 and -alpha7. PLoS Pathog. 2012, 8, e1002488. [Google Scholar] [CrossRef]
- Nitahara-Kasahara, Y.; Kamata, M.; Yamamoto, T.; Zhang, X.; Miyamoto, Y.; Muneta, K.; Iijima, S.; Yoneda, Y.; Tsunetsugu-Yokota, Y.; Aida, Y. Novel nuclear import of Vpr promoted by importin α is crucial for human immunodeficiency virus type 1 replication in macrophages. J. Virol. 2007, 81, 5284–5293. [Google Scholar] [CrossRef] [Green Version]
- Kuusisto, H.V.; Jans, D.A. Hyper-dependence of breast cancer cell types on the nuclear transporter Importin beta1. Biochim. Biophys. Acta 2015, 1853, 1870–1878. [Google Scholar] [CrossRef] [Green Version]
- Pumroy, R.A.; Ke, S.; Hart, D.J.; Zachariae, U.; Cingolani, G. Molecular determinants for nuclear import of influenza A PB2 by importin alpha isoforms 3 and 7. Structure 2015, 23, 374–384. [Google Scholar] [CrossRef]
- Guo, H.; Mao, R.; Block, T.M.; Guo, J.T. Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J. Virol. 2010, 84, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, M.; Saso, W.; Sugiyama, R.; Ishii, K.; Ohki, M.; Nagamori, S.; Suzuki, R.; Aizaki, H.; Ryo, A.; Yun, J.H.; et al. Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. Proc. Natl. Acad. Sci. USA 2019, 116, 8487–8492. [Google Scholar] [CrossRef]






| Gene | Forward Primer Sequence (5′ → 3′) | Reverse Primer Sequence (5′ → 3′) |
| HPRT | GCGATGTCAATAGGACTCCAG | TTGTTGTAGGATATGCCCTTGA |
| KPNA1 | GTGTCTGAATATCATCCCCTGTG | GAGACTTGTGGAACTGCTGAT |
| KPNA2 | ATCAACAGACCAATTTACAGTGC | AAGGATGACCAGATGCTGAAG |
| KPNA3 | CTGTTCATTACCTCCATCTGTCA | GCCAGCTTTATGTGTCCTCAT |
| KPNA4 | CCTTAATTCAACTACAACTTCATTTCG | CCAACGGCTCAAGAATTTCAAG |
| KPNA5 | TGATTGAAACTGGGGCTGT | GCATTCTGCATTGTCACCAG |
| KPNA6 | TCCATTACAGTCAGCAAGTCAC | CATCACCAATGCCACATCAG |
| KPNA7 | CCAGCTCAGAACTCAATGTCT | AGCACGTTCAGCATACCC |
| KPNB1 | ATTCTGCCTACAGTCCATGC | CCAGTCAGCTCAAACCACTA |
| Gene | Probe Sequence (5′ → 3′) | |
| HPRT | 56-FAM/AGCCTAAGA/ZEN/TGAGAGTTCAAGTTGAGTTTGG/31ABkFQ | |
| KPNA1 | 56-FAM/TTTCTCCTG/ZEN/CTTTGCGAGCTGTG/31ABkFQ | |
| KPNA2 | 56-FAM/TGATGATGC/ZEN/TACTTCTCCGCTGCAG/31ABkFQ | |
| KPNA3 | 56-FAM/ACAGAGCCC/ZEN/AAACAGTGTCTACAAGAATG/31ABkFQ | |
| KPNA4 | 56-FAM/TTCTCATAG/ZEN/TCTCCAAGTCGCGGC/31ABkFQ | |
| KPNA5 | 56-FAM/CAGGCTGTT/ZEN/TGGGCACTTGGTAAT/31ABkFQ | |
| KPNA6 | 56-FAM/TGATGCAGC/ZEN/CCAGTGAGACCAG/31ABkFQ | |
| KPNA7 | 56-FAM/ACAGATGAG/ZEN/CAGACGCAGATGGC/31ABkFQ | |
| KPNB1 | 56-FAM/TGCCCACCC/ZEN/TAATAGAATTAATGAAAGACCC/31ABkFQ | |
| Antibody Name | Source | Catalog Number | Biological Source | Dilution |
|---|---|---|---|---|
| KPNA1 | Proteintech | 18137-1-AP | Rabbit | 1:1000 |
| KPNA2 | GeneTex | GTX106323 | Rabbit | 1:1000 |
| KPNA3 | Bethyl Laboratories | A301-626A | Rabbit | 1:500 |
| KPNA4 | Proteintech | 12463-1-AP | Rabbit | 1:2000 |
| KPNA5 | GeneTex | GTX112203 | Rabbit | 1:2000 |
| KPNA6 | Proteintech | 12366-2-AP | Rabbit | 1:1000 |
| KPNB1 | Proteintech | 10077-1-AP | Rabbit | 1:3000 |
| Lamin B1 | Proteintech | 12987-1-AP | Rabbit | 1:2000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakanishi, A.; Okumura, H.; Hashita, T.; Yamashita, A.; Nishimura, Y.; Watanabe, C.; Kamimura, S.; Hayashi, S.; Murakami, S.; Ito, K.; et al. Ivermectin Inhibits HBV Entry into the Nucleus by Suppressing KPNA2. Viruses 2022, 14, 2468. https://doi.org/10.3390/v14112468
Nakanishi A, Okumura H, Hashita T, Yamashita A, Nishimura Y, Watanabe C, Kamimura S, Hayashi S, Murakami S, Ito K, et al. Ivermectin Inhibits HBV Entry into the Nucleus by Suppressing KPNA2. Viruses. 2022; 14(11):2468. https://doi.org/10.3390/v14112468
Chicago/Turabian StyleNakanishi, Anna, Hiroki Okumura, Tadahiro Hashita, Aya Yamashita, Yuka Nishimura, Chihiro Watanabe, Sakina Kamimura, Sanae Hayashi, Shuko Murakami, Kyoko Ito, and et al. 2022. "Ivermectin Inhibits HBV Entry into the Nucleus by Suppressing KPNA2" Viruses 14, no. 11: 2468. https://doi.org/10.3390/v14112468