Adenovirus 14p1 Immunopathogenesis during Lung Infection in the Syrian Hamster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Animals
2.3. Ethics Statement
2.4. Viral Infection of Animals
2.5. Histopathology of Lungs
2.6. Immunohistochemistry
2.7. Recovery of Immune Cells from Bronchoalveolar Lavage Fluid
2.8. Differential Cell Counts on BALF Cells
2.9. Quantitation of Ad14 Genomes in Hamster Lungs
2.10. Cytokine Gene Expression
2.11. Statistical Analysis
3. Results
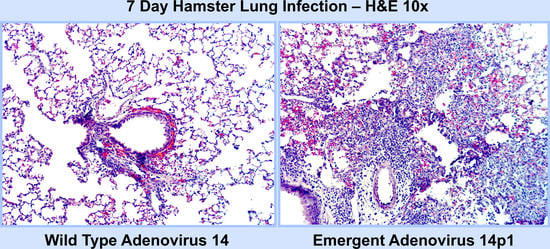
3.1. Ad14p1 Induces Severe Inflammation in the Lungs of Syrian Hamsters
3.2. Ad14 and Ad14p1 Induce a Mixed Leukocyte Immune Response
3.3. Ad14 and Ad14p1 Replicate in Syrian Hamsters
3.4. Ad14 and Ad14p1 Infections Are Associated with Different Cytokine/Chemokine Profiles
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van der Veen, J.; Kok, G. Isolation and typing of adenoviruses recovered from military recruits with acute respiratory disease in The Netherlands. Am. J. Epidemiol. 1957, 65, 119–129. [Google Scholar]
- Metzgar, D.; Osuna, M.; Kajon, A.E.; Hawksworth, A.W.; Irvine, M.; Russell, K.L. Abrupt emergence of diverse species B adenoviruses at US military recruit training centers. J. Infect. Dis. 2007, 196, 1465–1473. [Google Scholar] [CrossRef]
- Houng, H.S.H.; Gong, H.; Kajon, A.E.; Jones, M.S.; Kuschner, R.A.; Lyons, A.; Lott, L.; Lin, K.-H.; Metzgar, D. Genome sequences of human Adenovirus 14 isolates from mild respiratory cases and a fatal pneumonia, isolated during 2006-2007 epidemics in North America. Respir. Res. 2010, 11, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Jing, S.; Cheng, Z.; Yu, Z.; Dehghan, S.; Shamsaddini, A.; Yan, Y.; Li, M.; Seto, D. Comparative genomic analysis of two emergent human adenovirus type 14 respiratory pathogen isolates in China reveals similar yet divergent genomes. Emerg. Microbes Infect. 2017, 6, e92-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, Z.; Butt, A.M.; An, X.; Jiang, T.; Liu, W.; Qin, C.; Cao, W.C.; Tong, Y. Genomic analysis of HAdV-B14 isolate from the outbreak of febrile respiratory infection in China. Genomics 2013, 102, 448–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, L.; An, J.; Xie, Z.; Dehghan, S.; Seto, D.; Xu, W.; Ji, Y. Genome and bioinformatic analysis of a HAdV-B14p1 virus isolated from a baby with pneumonia in Beijing, China. PLoS ONE 2013, 8, e60345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, M.J.; Kajon, A.E.; Lu, X.; Dunford, L.; O’Reilly, P.; Holder, P.; De Gascun, C.F.; Coughlan, S.; Connell, J.; Erdman, D.D.; et al. Deaths associated with human adenovirus-14p1 infections, Europe, 2009–2010. Emerg. Infect. Dis. 2011, 17, 1402–1408. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.F.; Schmidt, M.A.; Lu, X.; Erdman, D.D.; Campbell, M.; Thomas, A.; Cieslak, P.R.; Grenz, L.D.; Tsaknardis, L.; Gleaves, C.; et al. A community-based outbreak of severe respiratory illness caused by human adenovirus serotype 14. J. Infect. Dis. 2009, 199, 1427–1434. [Google Scholar] [CrossRef] [Green Version]
- Louie, J.K.; Kajon, A.E.; Holodniy, M.; Guardia-LaBar, L.; Lee, B.; Petru, A.M.; Hacker, J.K.; Schnurr, D.P. Severe pneumonia due to adenovirus serotype 14: A new respiratory threat? Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2008, 46, 421–425. [Google Scholar] [CrossRef]
- Tate, J.E.; Bunning, M.L.; Lott, L.; Lu, X.; Su, J.; Metzgar, D.; Brosch, L.; Panozzo, C.A.; Marconi, V.C.; Faix, D.J.; et al. Outbreak of severe respiratory disease associated with emergent human adenovirus serotype 14 at a US air force training facility in 2007. J. Infect. Dis. 2009, 199, 1419–1426. [Google Scholar] [CrossRef] [Green Version]
- O’Flanagan, D.; O’Donnell, J.; Domegan, L.; Fitzpatrick, F.; Connell, J.; Coughlan, S.; De Gascun, C.; Carr, M.J. First reported cases of human adenovirus serotype 14p1 infection, Ireland, October 2009 to July 2010. Euro. Surveill. 2011, 16, 19801. [Google Scholar]
- Esposito, D.H.; Gardner, T.J.; Schneider, E.; Stockman, L.J.; Tate, J.E.; Panozzo, C.A.; Robbins, C.L.; Jenkerson, S.A.; Thomas, L.; Watson, C.M.; et al. Outbreak of pneumonia associated with emergent human adenovirus serotype 14--Southeast Alaska, 2008. J. Infect. Dis. 2010, 202, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Kajon, A.E.; Lu, X.; Erdman, D.D.; Louie, J.; Schnurr, D.; George, K.S.; Koopmans, M.P.; Allibhai, T.; Metzgar, D. Molecular epidemiology and brief history of emerging adenovirus 14-associated respiratory disease in the United States. J. Infect. Dis. 2010, 202, 93–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girouard, G.; Garceau, R.; Thibault, L.; Oussedik, Y.; Bastien, N.; Li, Y. Adenovirus serotype 14 infection, New Brunswick, Canada, 2011. Emerg. Infect. Dis. 2013, 19, 119–122. [Google Scholar] [CrossRef]
- Anderson, B.D.; Barr, K.L.; Heil, G.L.; Friary, J.A.; Gray, G.C. A comparison of viral fitness and virulence between emergent adenovirus 14p1 and prototype adenovirus 14p strains. J. Clin. Virol. 2012, 54, 265–268. [Google Scholar] [CrossRef] [Green Version]
- Lam, E.; Ramke, M.; Warnecke, G.; Schrepfer, S.; Kopfnagel, V.; Dobner, T.; Heim, A. Effective Apical Infection of Differentiated Human Bronchial Epithelial Cells and Induction of Proinflammatory Chemokines by the Highly Pneumotropic Human Adenovirus Type 14p1. PLoS ONE 2015, 10, e0131201. [Google Scholar] [CrossRef] [Green Version]
- Radke, J.R.; Grigera, F.; Ucker, D.S.; Cook, J.L. Adenovirus E1B 19-kilodalton protein modulates innate immunity through apoptotic mimicry. J. Virol. 2014, 88, 2658–2669. [Google Scholar] [CrossRef] [Green Version]
- Radke, J.R.; Yong, S.L.; Cook, J.L. Low-Level Expression of the E1B 20-Kilodalton Protein by Adenovirus 14p1 Enhances Viral Immunopathogenesis. J. Virol. 2016, 90, 497–505. [Google Scholar] [CrossRef] [Green Version]
- Stewart, S.; Fishbein, M.C.; Snell, G.I.; Berry, G.J.; Boehler, A.; Burke, M.M.; Glanville, A.; Gould, F.K.; Magro, C.; Marboe, C.C.; et al. Revision of the 1996 working formulation for the standardization of nomenclature in the diagnosis of lung rejection. J. Heart Lung Transplant. 2007, 26, 1229–1242. [Google Scholar] [CrossRef]
- Zivcec, M.; Safronetz, D.; Haddock, E.; Feldmann, H.; Ebihara, H. Validation of assays to monitor immune responses in the Syrian golden hamster (Mesocricetus auratus). J. Immunol. Methods 2011, 368, 24–35. [Google Scholar] [CrossRef]
- Zhang, Q.; Seto, D.; Zhao, S.; Zhu, L.; Zhao, W.; Wan, C. Genome Sequence of the First Human Adenovirus Type 14 Isolated in China. J. Virol. 2012, 86, 7019–7020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parcell, B.J.; McIntyre, P.G.; Yirrell, D.L.; Fraser, A.; Quinn, M.; Templeton, K.; Christie, S.; Romanes, F. Prison and community outbreak of severe respiratory infection due to adenovirus type 14p1 in Tayside, UK. J. Public Health (Oxf.) 2015, 37, 64–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Yu, D.; Zhu, Z.; Zhao, H.; Wang, P.; Gray, G.C.; Meng, L.; Xu, W. Outbreak of febrile respiratory illness associated with human adenovirus type 14p1 in Gansu Province, China. Influenza Other Respi. Viruses 2013, 7, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Ying, B.; Toth, K.; Spencer, J.F.; Aurora, R.; Wold, W.S.M. Transcriptome sequencing and development of an expression microarray platform for liver infection in adenovirus type 5-infected Syrian golden hamsters. Virology 2015, 485, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.A.; Spencer, J.F.; La Regina, M.C.; Dhar, D.; Tollefson, A.E.; Toth, K.; Wold, W.S.M. Syrian hamster as a permissive immunocompetent animal model for the study of oncolytic adenovirus vectors. Cancer Res. 2006, 66, 1270–1276. [Google Scholar] [CrossRef] [Green Version]
- Tollefson, A.E.; Ying, B.; Spencer, J.F.; Sagartz, J.E.; Wold, W.S.M.; Toth, K. Pathology in Permissive Syrian Hamsters after Infection with Species C Human Adenovirus (HAdV-C) Is the Result of Virus Replication: HAdV-C6 Replicates More and Causes More Pathology than HAdV-C5. J. Virol. 2017, 91, e00284-17. [Google Scholar] [CrossRef] [Green Version]
- Prince, G.A.; Porter, D.D.; Jenson, A.B.; Horswood, R.L.; Chanock, R.M.; Ginsberg, H.S. Pathogenesis of adenovirus type 5 pneumonia in cotton rats (Sigmodon hispidus). J. Virol. 1993, 67, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Yei, S.; Mittereder, N.; Wert, S.; Whitsett, J.A.; Wilmott, R.W.; Trapnell, B.C. In vivo evaluation of the safety of adenovirus-mediated transfer of the human cystic fibrosis transmembrane conductance regulator cDNA to the lung. Hum. Gene Ther. 1994, 5, 731–744. [Google Scholar] [CrossRef]
- Ginsberg, H.S.; Moldawer, L.L.; Prince, G.A. Role of the type 5 adenovirus gene encoding the early region 1B 55-kDa protein in pulmonary pathogenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 10409–10411. [Google Scholar] [CrossRef] [Green Version]
- McCoy, R.D.; Davidson, B.L.; Roessler, B.J.; Huffnagle, G.B.; Janich, S.L.; Laing, T.J.; Simon, R.H. Pulmonary inflammation induced by incomplete or inactivated adenoviral particles. Hum. Gene Ther. 1995, 6, 1553–1560. [Google Scholar] [CrossRef] [Green Version]
- Otake, K.; Ennist, D.L.; Harrod, K.; Trapnell, B.C. Nonspecific inflammation inhibits adenovirus-mediated pulmonary gene transfer and expression independent of specific acquired immune responses. Hum. Gene Ther. 1998, 9, 2207–2222. [Google Scholar] [CrossRef] [PubMed]
- Maler, M.D.; Nielsen, P.J.; Stichling, N.; Cohen, I.; Ruzsics, Z.; Wood, C.; Engelhard, P.; Suomalainen, M.; Gyory, I.; Huber, M. Key Role of the Scavenger Receptor MARCO in Mediating Adenovirus Infection and Subsequent Innate Responses of Macrophages. MBio 2017, 8, 855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stichling, N.; Suomalainen, M.; Flatt, J.W.; Schmid, M.; Pacesa, M.; Hemmi, S.; Jungraithmayr, W.; Maler, M.D.; Freudenberg, M.A.; Plückthun, A.; et al. Lung macrophage scavenger receptor SR-A6 (MARCO) is an adenovirus type-specific virus entry receptor. PLoS Pathog. 2018, 14, e1006914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appledorn, D.M.; Patial, S.; McBride, A.; Godbehere, S.; Van Rooijen, N.; Parameswaran, N.; Amalfitano, A. Adenovirus vector-induced innate inflammatory mediators, MAPK signaling, as well as adaptive immune responses are dependent upon both TLR2 and TLR9 in vivo. J. Immunol. 2008, 181, 2134–2144. [Google Scholar] [CrossRef]
- Zhou, X.; Ramke, M.; Chintakuntlawar, A.V.; Lee, J.Y.; Rajaiya, J.; Chodosh, J. Role of MyD88 in adenovirus keratitis. Immunol. Cell Biol. 2017, 95, 108–116. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Li, Z.-Y.; Liu, Y.; Persson, J.; Beyer, I.; Möller, T.; Koyuncu, D.; Drescher, M.R.; Strauss, R.; Zhang, X.-B.; et al. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat. Med. 2010, 17, 96–104. [Google Scholar] [CrossRef]
- Wang, H.; Tuve, S.; Erdman, D.D.; Lieber, A. Receptor usage of a newly emergent adenovirus type 14. Virology 2009, 387, 436–441. [Google Scholar] [CrossRef]
- Lavery, D.; Fu, S.M.; Lufkin, T.; Chen-Kiang, S. Productive infection of cultured human lymphoid cells by adenovirus. J. Virol. 1987, 61, 1466–1472. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Gibbons, J.G.; DeLoid, G.M.; Bedugnis, A.S.; Thimmulappa, R.K.; Biswal, S.; Kobzik, L. Immunomodulators targeting MARCO expression improve resistance to postinfluenza bacterial pneumonia. Am. J. Physiol. Lung Cell Mol. Physiol. 2017, 313, L138–L153. [Google Scholar] [CrossRef]
- ARDS Definition Task Force; Ranieri, V.M.; Rubenfeld, G.D.; Thompson, B.T.; Ferguson, N.D.; Caldwell, E.; Fan, E.; Camporota, L.; Slutsky, A.S. Acute respiratory distress syndrome: The Berlin Definition. J. Am. Med. Assoc. 2012, 307, 2526–2533. [Google Scholar]
- Matute-Bello, G.; Downey, G.; Moore, B.B.; Groshong, S.D.; Matthay, M.A.; Slutsky, A.S.; Kuebler, W.M. Acute Lung Injury in Animals Study Group An official American Thoracic Society workshop report: Features and measurements of experimental acute lung injury in animals. Am. J. Respir. Cell Mol. Biol. 2011, 44, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aeffner, F.; Bolon, B.; Davis, I.C. Mouse Models of Acute Respiratory Distress Syndrome: A Review of Analytical Approaches, Pathologic Features, and Common Measurements. Toxicol. Pathol. 2015, 43, 1074–1092. [Google Scholar] [CrossRef] [PubMed]
- Uhlig, S.; Kuebler, W.M. Difficulties in modelling ARDS (2017 Grover Conference Series). Pulm. Circ. 2018, 8, 2045894018766737. [Google Scholar] [CrossRef] [PubMed] [Green Version]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radke, J.R.; Covert, H.J.; Bauer, F.; Ananthanarayanan, V.; Cook, J.L. Adenovirus 14p1 Immunopathogenesis during Lung Infection in the Syrian Hamster. Viruses 2020, 12, 595. https://doi.org/10.3390/v12060595
Radke JR, Covert HJ, Bauer F, Ananthanarayanan V, Cook JL. Adenovirus 14p1 Immunopathogenesis during Lung Infection in the Syrian Hamster. Viruses. 2020; 12(6):595. https://doi.org/10.3390/v12060595
Chicago/Turabian StyleRadke, Jay R., Hunter J. Covert, Fredrick Bauer, Vijayalakshmi Ananthanarayanan, and James L. Cook. 2020. "Adenovirus 14p1 Immunopathogenesis during Lung Infection in the Syrian Hamster" Viruses 12, no. 6: 595. https://doi.org/10.3390/v12060595