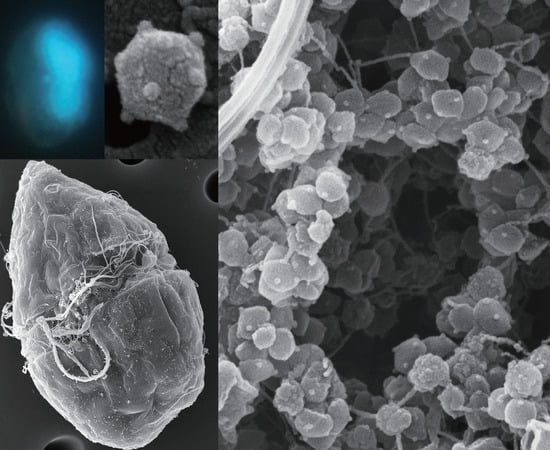
Visualization of a Dinoflagellate-Infecting Virus HcDNAV and Its Infection Process
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Falkowski, P.G.; Katz, M.E.; Knoll, A.H.; Quigg, A.; Raven, J.A.; Schofield, O.; Taylor, F.J. The evolution of modern eukaryotic phytoplankton. Science 2004, 305, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Hallegraeff, G.M. A review of harmful algal blooms and their apparent global increase. Phycologia 1993, 32, 79–99. [Google Scholar] [CrossRef]
- Erdner, D.L.; Dyble, J.; Parsons, M.L.; Stevens, R.C.; Hubbard, K.A.; Wrabel, M.L.; Moore, S.K.; Lefebvre, K.A.; Anderson, D.M.; Bienfang, P.; et al. Centers for Oceans and Human Health: A unified approach to the challenge of harmful algal blooms. Environ. Health 2008, 7 (Suppl. 2), S2. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, T. Heterocapsa circularisquama sp. nov. (Peridiniales, Dinophyceae): A new marine dinoflagellate causing mass mortality of bivalves in Japan. Phycol. Res. 1995, 43, 129–136. [Google Scholar] [CrossRef]
- Matsuyama, Y. Harmful effect of dinoflagellate Heterocapsa circularisquama on shellfish aquaculture in Japan. Jpn. Agric. Res. Q. 1999, 33, 283–293. [Google Scholar]
- Tomaru, Y.; Katanozaka, N.; Nishida, K.; Shirai, Y.; Tarutani, K.; Yamaguchi, M.; Nagasaki, K. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat. Microb. Ecol. 2004, 34, 207–218. [Google Scholar] [CrossRef]
- Claverie, J.M.; Ogata, H.; Audic, S.; Abergel, C.; Suhre, K.; Fournier, P.E. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006, 117, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claverie, J.M.; Ogata, H. Ten good reasons not to exclude giruses from the evolutionary picture. Nat. Rev. Microbiol. 2009, 7, 615. [Google Scholar] [CrossRef] [PubMed]
- Tarutani, K.; Nagasaki, K.; Itakura, S.; Yamaguchi, M. Isolation of a virus infecting the novel shellfish-killing dinoflagellate Heterocapsa circularisquama. Aquat Microb. Ecol. 2001, 23, 103–111. [Google Scholar] [CrossRef]
- Nagasaki, K.; Tomaru, Y.; Tarutani, K.; Katanozaka, N.; Yamanaka, S.; Tanabe, H.; Yamaguchi, M. Growth characteristics and intraspecies host specificity of a large virus infecting the dinoflagellate Heterocapsa circularisquama. Appl. Environ. Microbiol. 2003, 69, 2580–2586. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki, K. Dinoflagellates, diatoms, and their viruses. J. Microbiol. 2008, 46, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Tomaru, Y.; Nagasaki, K. Widespread occurrence of viruses lytic to the bivalve-killing dinoflagellate Heterocapsa circularisquama along the western coast of Japan. Plankton Biol. Ecol. 2004, 51, 1–6. [Google Scholar]
- Nagasaki, K.; Shirai, Y.; Tomaru, Y.; Nishida, K.; Pietrokovski, S. Algal viruses with distinct intraspecies host specificities include identical intein elements. Appl. Environ. Microbiol. 2005, 71, 3599–3607. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Roznowski, A.P.; Tokuda, J.M.; Klose, T.; Mauney, A.; Pollack, L.; Fane, B.A.; Rossmann, M.G. Structural changes of tailless bacteriophage PhiX174 during penetration of bacterial cell walls. Proc. Natl. Acad. Sci. USA 2017, 114, 13708–13713. [Google Scholar] [CrossRef] [PubMed]
- Agarkova, I.; Hertel, B.; Zhang, X.; Lane, L.; Tchourbanov, A.; Dunigan, D.D.; Thiel, G.; Rossmann, M.G.; Van Etten, J.L. Dynamic attachment of Chlorovirus PBCV-1 to Chlorella variabilis. Virology 2014, 466–467, 95–102. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takano, Y.; Tomaru, Y.; Nagasaki, K. Visualization of a Dinoflagellate-Infecting Virus HcDNAV and Its Infection Process. Viruses 2018, 10, 554. https://doi.org/10.3390/v10100554
Takano Y, Tomaru Y, Nagasaki K. Visualization of a Dinoflagellate-Infecting Virus HcDNAV and Its Infection Process. Viruses. 2018; 10(10):554. https://doi.org/10.3390/v10100554
Chicago/Turabian StyleTakano, Yoshihito, Yuji Tomaru, and Keizo Nagasaki. 2018. "Visualization of a Dinoflagellate-Infecting Virus HcDNAV and Its Infection Process" Viruses 10, no. 10: 554. https://doi.org/10.3390/v10100554