Analysis of Plant Trait Data of Host Plants of Lycorma delicatula in the US Suggests Evidence for Ecological Fitting
Abstract
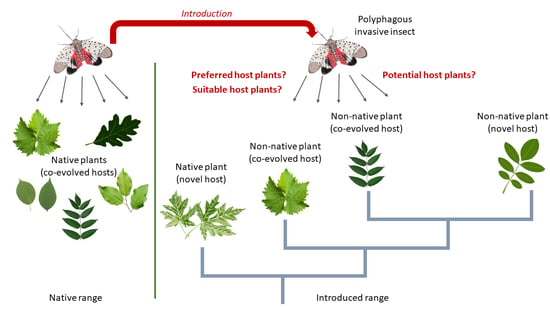
:1. Introduction
2. Materials and Methods
2.1. Plant Trait Data Collection and Processing
2.2. Plant Trait Data Synthesis and Statistical Analysis
2.3. Phylogenetic Reconstruction and Genetic Distance Analysis
3. Results
3.1. Plant Trait Data and Environmental Requirements
3.2. Phylogenetic Reconstruction and Genetic Distance from Ailanthus altissima
4. Discussion
4.1. Plant Trait Data and Environmental Requirements
4.2. Plant Phylogenetic Relatedness and Ecological Fitting
4.3. Potential Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruesink, J.L.; Parker, I.M.; Groom, M.J.; Kareiva, P.M. Reducing the risks of non-indigenous species introductions. BioScience 1995, 45, 465–477. [Google Scholar] [CrossRef]
- Mazzi, D.; Dorn, S. Movement of insect pests in agricultural landscapes. Ann. Appl. Biol. 2012, 160, 97–113. [Google Scholar] [CrossRef]
- Schindler, S.; Staska, B.; Adam, M.; Rabitsch, W.; Essl, F. Alien species and public health impacts in Europe: A literature review. NeoBiota 2015, 27, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Simberloff, D.; Martin, J.-L.; Genovesi, P.; Maris, V.; Wardle, D.A.; Aronson, J.; Courchamp, F.; Galil, B.; García-Berthou, E.; Pascal, M. Impacts of biological invasions: What’s what and the way forward. Trends Ecol. Evol. 2013, 28, 58–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimentel, D.; Zuniga, R.; Morrison, D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 2005, 52, 273–288. [Google Scholar] [CrossRef]
- Alpert, P. The advantages and disadvantages of being introduced. Biol. Invasions 2006, 8, 1523–1534. [Google Scholar] [CrossRef]
- Pyšek, P.; Jarošík, V.; Hulme, P.E.; Pergl, J.; Hejda, M.; Schaffner, U.; Vilà, M. A global assessment of invasive plant impacts on resident species, communities and ecosystems: The interaction of impact measures, invading species’ traits and environment. Glob. Chang. Biol. 2012, 18, 1725–1737. [Google Scholar] [CrossRef]
- Liebhold, A.M.; Yamanaka, T.; Roques, A.; Augustin, S.; Chown, S.L.; Brockerhoff, E.G.; Pyšek, P. Global compositional variation among native and non-native regional insect assemblages emphasizes the importance of pathways. Biol. Invasions 2016, 18, 893–905. [Google Scholar] [CrossRef] [Green Version]
- Sax, D.F.; Stachowicz, J.J.; Brown, J.H.; Bruno, J.F.; Dawson, M.N.; Gaines, S.D.; Grosberg, R.K.; Hastings, A.; Holt, R.D.; Mayfield, M.M.; et al. Ecological and evolutionary insights from species invasions. Trends Ecol. Evol. 2007, 22, 465–471. [Google Scholar] [CrossRef]
- Agosta, S.J.; Klemens, J.A. Ecological fitting by phenotypically flexible genotypes: Implications for species associations, community assembly and evolution. Ecol. Lett. 2008, 11, 1123–1134. [Google Scholar] [CrossRef]
- Janzen, D.H. On ecological fitting. Oikos 1985, 45, 308–310. [Google Scholar] [CrossRef] [Green Version]
- Agosta, S.J. On ecological fitting, plant–insect associations, herbivore host shifts, and host plant selection. Oikos 2006, 114, 556–565. [Google Scholar] [CrossRef]
- Agosta, S.J.; Klemens, J.A. Resource specialization in a phytophagous insect: No evidence for genetically based performance trade-offs across hosts in the field or laboratory. J. Evol. Biol. 2009, 22, 907–912. [Google Scholar] [CrossRef]
- Brooks, D.R.; León-Règagnon, V.; McLennan, D.A.; Zelmer, D. Ecological fitting as a determinant of the community structure of platyhelminth parasites of anurans. Ecology 2006, 87 (Suppl. S7), S76–S85. [Google Scholar] [CrossRef]
- Kattge, J.; Bönisch, G.; Díaz, S.; Lavorel, S.; Prentice, I.C.; Leadley, P.; Tautenhahn, S.; Werner, G.D.A.; Aakala, T.; Abedi, M.; et al. TRY plant trait database–enhanced coverage and open access. Glob. Chang. Biol. 2019, 26, 119–188. [Google Scholar] [CrossRef] [Green Version]
- Cipollini, D.; Peterson, D.L. The potential for host switching via ecological fitting in the emerald ash borer-host plant system. Oecologia 2018, 187, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Gougherty, A.V.; Davies, T.J. Towards a phylogenetic ecology of plant pests and pathogens. Philos. Trans. R. Soc. B 2021, 376, 20200359. [Google Scholar] [CrossRef]
- National Invasive Species Information Center; U.S. Department of Agriculture. Executive Order 13112 of February 3, 1999; Section 1. Available online: https://www.invasivespeciesinfo.gov/executive-order-13112 (accessed on 2 August 2022).
- Saul, W.-C.; Jeschke, J.M. Eco-evolutionary experience in novel species interactions. Ecol. Lett. 2015, 18, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Avanesyan, A.; Lamp, W.O. Use of molecular gut content analysis to decipher the range of food plants of the invasive spotted lanternfly, Lycorma delicatula. Insects 2020, 11, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McPherson, C.; Avanesyan, A.; Lamp, W.O. Diverse Host Plants of the First Instars of the Invasive Lycorma delicatula: Insights from eDNA Metabarcoding. Insects 2022, 13, 534. [Google Scholar] [CrossRef]
- Tabashnik, B.E. Host range evolution: The shift from native legume hosts to alfalfa by the butterfly, Colias philodice eriphyle. Evolution 1983, 37, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Janzen, D.H. When is it coevolution? Evolution 1980, 34, 611–612. [Google Scholar] [CrossRef] [PubMed]
- Barringer, L.; Ciafré, C.M. Worldwide feeding host plants of spotted lanternfly, with significant additions from North America. Environ. Entomol. 2020, 49, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: http://www.R-project.org/ (accessed on 2 August 2022).
- Rausher, M.D. Natural selection and the evolution of plant-insect interactions. In Insect Chemical Ecology: An Evolutionary Approach; Chapman and Hall: New York, NY, USA, 1992; pp. 20–88. [Google Scholar]
- Price, P.W.; Denno, R.F.; Eubanks, M.D.; Finke, D.L.; Kaplan, I. Insect Ecology: Behavior, Populations and Communities; Cambridge University Press: Cambridge, England, 2011. [Google Scholar]
- Kim, J.G.; Lee, E.-H.; Seo, Y.-M.; Kim, N.-Y. Cyclic Behavior of Lycorma delicatula (Insecta: Hemiptera: Fulgoridae) on Host Plants. J. Insect Behav. 2011, 24, 423–435. [Google Scholar] [CrossRef]
- Pearse, I.S.; Hipp, A.L. Native plant diversity increases herbivory to non-natives. Proc. R. Soc. B Biol. Sci. 2014, 281, 20141841. [Google Scholar] [CrossRef]
- Dara, S.K.; Barringer, L.; Arthurs, S.P. Lycorma delicatula (Hemiptera: Fulgoridae): A new invasive pest in the United States. J. Integr. Pest Manag. 2015, 6, 20. [Google Scholar] [CrossRef] [Green Version]
- Clifton, E.H.; Hajek, A.E.; Jenkins, N.E.; Roush, R.T.; Rost, J.P.; Biddinger, D.J. Applications of Beauveria bassiana (Hypocreales: Cordycipitaceae) to Control Populations of Spotted Lanternfly (Hemiptera: Fulgoridae), in Semi-Natural Landscapes and on Grapevines. Environ. Entomol. 2020, 49, 854–864. [Google Scholar] [CrossRef]
- Nixon, L.J.; Jones, S.; Dechaine, A.C.; Ludwick, D.; Hickin, M.; Sullivan, L.; Elsensohn, J.E.; Gould, J.; Keena, M.; Kuhar, T.; et al. Development of rearing methodology for the invasive Spotted Lanternfly, Lycorma delicatula (Hemiptera: Fulgoridae). Front. Insect Sci. 2022, 2, 1025193. [Google Scholar] [CrossRef]
- Hanan, J.; Prusinkiewicz, P.; Zalucki, M.; Skirvin, D. Simulation of insect movement with respect to plant architecture and morphogenesis. Comput. Electron. Agric. 2002, 35, 255–269. [Google Scholar] [CrossRef]
- Kane, S.A.; Bien, T.; Contreras-Orendain, L.; Ochs, M.F.; Tonia Hsieh, S. Many ways to land upright: Novel righting strategies allow spotted lanternfly nymphs to land on diverse substrates. J. R. Soc. Interface 2021, 18, 20210367. [Google Scholar] [CrossRef]
- Price, P.W.; Andrade, I.; Pires, C.; Sujii, E.; Vieira, E.M. Gradient Analysis Using Plant Modular Structure: Pattern in Plant Architecture and Insect Herbivore Utilization. Environ. Entomol. 1995, 24, 497–505. [Google Scholar] [CrossRef]
- Wolfin, M.S.; Binyameen, M.; Wang, Y.; Urban, J.M.; Roberts, D.C.; Baker, T.C. Flight Dispersal Capabilities of Female Spotted Lanternflies (Lycorma delicatula) Related to Size and Mating Status. J. Insect Behav. 2019, 32, 188–200. [Google Scholar] [CrossRef]
- Liu, H.; Lusk, R.; Gallardy, R. Infrared thermography for insect detection: Lighting up the spotted lanternfly in the field. J. Pest Sci. 2021, 94, 231–240. [Google Scholar] [CrossRef]
- Grevstad, F.S.; Klepetka, B.W. The influence of plant architecture on the foraging efficiencies of a suite of ladybird beetles feeding on aphids. Oecologia 1992, 92, 399–404. [Google Scholar] [CrossRef]
- Jung, J.-M.; Jung, S.; Byeon, D.-H.; Lee, W.-H. Model-based prediction of potential distribution of the invasive insect pest, spotted lanternfly Lycorma delicatula (Hemiptera: Fulgoridae), by using CLIMEX. J. Asia-Pac. Biodivers. 2017, 10, 532–538. [Google Scholar] [CrossRef]
- Jones, C.; Skrip, M.M.; Seliger, B.J.; Jones, S.; Wakie, T.; Takeuchi, Y.; Petras, V.; Petrasova, A.; Meentemeyer, R.K. Spotted lanternfly predicted to establish in California by 2033 without preventative management. Commun. Biol. 2022, 5, 558. [Google Scholar] [CrossRef] [PubMed]
- Smyers, E.C.; Urban, J.M.; Dechaine, A.C.; Pfeiffer, D.G.; Crawford, S.R.; Calvin, D.D. Spatio-Temporal Model for Predicting Spring Hatch of the Spotted Lanternfly (Hemiptera: Fulgoridae). Environ. Entomol. 2020, 50, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Rasmann, S.; Agrawal, A.A. Evolution of Specialization: A Phylogenetic Study of Host Range in the Red Milkweed Beetle (Tetraopes tetraophthalmus). Am. Nat. 2011, 177, 728–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, G.S.; Briggs, H.M.; Magarey, R. The Impact of Plant Enemies Shows a Phylogenetic Signal. PLoS ONE 2015, 10, e0123758. [Google Scholar] [CrossRef]
- Murphy, S.M.; Feeny, P. Chemical facilitation of a naturally occurring host shift by Papilio machaon butterflies (Papilionidae). Ecol. Monogr. 2006, 76, 399–414. [Google Scholar] [CrossRef]
- Pearse, I.S.; Hipp, A.L. Phylogenetic and trait similarity to a native species predict herbivory on non-native oaks. Proc. Natl. Acad. Sci. USA 2009, 106, 18097–18102. [Google Scholar] [CrossRef] [Green Version]
- Avanesyan, A.; Culley, T.M. Feeding preferences of Melanoplus femurrubrum grasshoppers on native and exotic grasses: Behavioral and molecular approaches. Entomol. Exp. Appl. 2015, 157, 152–163. [Google Scholar] [CrossRef]
- Rahmathulla, V.K.; Kumar, C.M.K.; Angadi, B.S.; Sivaprasad, V. Association of Climatic Factors on Population Dynamics of Leaf Roller, Diaphania pulverulentalis Hampson (Lepidoptera: Pyralidae) in Mulberry Plantations of Sericulture Seed Farm. Psyche A J. Entomol. 2012, 2012, 186214. [Google Scholar] [CrossRef] [Green Version]
- Bălăcenoiu, F.; Simon, D.C.; Nețoiu, C.; Toma, D.; Petrițan, I.C. The Seasonal Population Dynamics of Corythucha arcuata (Say, 1832) (Hemiptera: Tingidae) and the Relationship between Meteorological Factors and the Diurnal Flight Intensity of the Adults in Romanian Oak Forests. Forests 2021, 12, 1774. [Google Scholar] [CrossRef]
- Sharma, D.; Maqbool, A.; Ahmad, H.; Srivastava, K.; Kumar, M.; Vir, V.; Jamwal, S. Effect of meteorological factors on the population dynamics of insect pests of tomato. Veg. Sci. 2013, 40, 90–92. [Google Scholar]
- Williams, C.B. Studies in the effect of weather conditions on the activity and abundance of insect populations. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1961, 244, 331–378. [Google Scholar]





| Term | Definition |
|---|---|
| Native species | “a species that, other than as a result of an introduction, historically occurred or currently occurs in that ecosystem” [18]. |
| Invasive species | A non-native species in relation to a particular ecosystem, “whose introduction causes or is likely to cause economic or environmental harm or harm to human health” [18]. |
| Novel species association | The association between resident (e.g., native plant) and non-resident (e.g., introduced insect) species, “in which at least one species has little or no experience with relevant ecological traits of its interaction counterpart” [19]. |
| Host plants | Following our previous research on host plant usage by L. delicatula [20,21], we will use the term “host plants” to refer to insect food plants—i.e., the plants which an insect pest utilizes as a food source and not for resting, molting, egg-laying, etc. |
| Host plant range | For the purpose of this proposed study, we will use the term “host plant range” to indicate the range of plants on which an insect pest feeds. Insect pests with a wide host plant range are polyphagous. |
| Host plant shift | A process “by which one or more formerly used host plant species are abandoned in favor of one or more new host plant species” [22]. |
| Host plant range expansion | A process of “the addition of one or more host plant species to the total number used by the herbivore species” [22]. |
| Ecological fitting | “the process whereby organisms colonize and persist in novel environments, use novel resources or form novel associations with other species as a result of the suites of traits that they carry at the time they encounter the novel condition” [10]. |
| Coevolution | “an evolutionary change in a trait of the individuals in one population in response to a trait of the individuals of a second population, followed by an evolutionary response by the second population to the change in the first” [23]. For the purpose of this proposed study, we consider such evolutionary change in a trait of a certain native plant species in response to herbivory by a certain native insect species, and vice versa (i.e., adaptations of certain native insects to feeding on certain native plants). |
| # 1 | Family 2 | Genus 2 | Species 2 | Common Name | Plant Origin 3 | Life Form 3 | Perenniality 3 |
|---|---|---|---|---|---|---|---|
| 1 | Aceraceae | Acer | buergerianum | Trident maple | Introduced | Tree | Perennial |
| 2 | Aceraceae | Acer | negundo | Boxelder | Native | Tree | Perennial |
| 3 | Aceraceae | Acer | palmatum | Japanese maple | Introduced | Shrub/tree | Perennial |
| 4 | Aceraceae | Acer | pictum | Yellow-paint maple | Introduced | Tree | Perennial |
| 5 | Aceraceae | Acer | platanoides | Norway maple | Introduced | Tree | Perennial |
| 6 | Aceraceae | Acer | pseudoplatanus | Sycamore maple | Introduced | Tree | Perennial |
| 7 | Aceraceae | Acer | rubrum | Red maple | Native | Tree | Perennial |
| 8 | Aceraceae | Acer | saccharinum | Silver maple | Native | Tree | Perennial |
| 9 | Simaroubaceae | Ailanthus | altissima | Tree of heaven | Introduced | Tree | Perennial |
| 10 | Asteraceae | Arctium | lappa | Greater burdock | Introduced | Forb/Herb | Biennial |
| 11 | Brassicaceae | Armoracia | rusticana | Horseradish | Introduced | Forb/Herb | Perennial |
| 12 | Betulaceae | Betula | pendula | European white birch | Introduced | Tree | Perennial |
| 13 | Betulaceae | Betula | alleghaniensis | Yellow birch | Native | Tree | Perennial |
| 14 | Betulaceae | Betula | lenta | Sweet birch | Native | Tree | Perennial |
| 15 | Celastraceae | Celastrus | orbiculatus | Oriental bittersweet | Introduced | Woody vine | Perennial |
| 16 | Fagaceae | Fagus | grandifolia | American beech | Native | Tree | Perennial |
| 17 | Cannabaceae | Humulus | lupulus | Common hop | Both | Forb/Herb/Vine | Perennial |
| 18 | Juglandaceae | Juglans | cinerea | Butternut | Native | Tree | Perennial |
| 19 | Juglandaceae | Juglans | nigra | Black walnut | Native | Tree | Perennial |
| 20 | Magnoliaceae | Liriodendron | tulipfera | Tulip tree | Native | Tree | Perennial |
| 21 | Fabaceae | Maackia | amurensis | Amur maackia | Introduced | Tree | Perennial |
| 22 | Meliaceae | Melia | azedarach | Chinaberrytree | Native | Shrub/Tree | Perennial |
| 23 | Cornaceae | Nyssa | sylvatica | blackgum | Native | Tree | Perennial |
| 24 | Vitaceae | Parthenocissus | quinquefolia | Virginia creeper | Native | Woody vine | Perennial |
| 25 | Rosaceae | Prunus | serotina | Black cherry | Native | Shrub/Tree | Perennial |
| 26 | Fagaceae | Quercus | acutissima | Sawtooth oak | Introduced | Tree | Perennial |
| 27 | Fagaceae | Quercus | rubra | Northern red oak | Native | Tree | Perennial |
| 28 | Anacardiaceae | Rhus | typhina | Staghorn sumac | Native | Shrub/Tree | Perennial |
| 29 | Fabaceae | Robinia | pseudoacacia | Black locust | Native | Tree | Perennial |
| 30 | Salicaceae | Salix | udensis | Japanese fantail willow | Introduced | Tree | Perennial |
| 31 | Salicaceae | Salix | babylonica | Weeping willow | Introduced | Tree | Perennial |
| 32 | Styracaceae | Styrax | japonicus | Japanese snowbell | Introduced | Shrub/Tree | Perennial |
| 33 | Cupressaceae | Thuja | occidentalis | Arborvitae | Native | Tree | Perennial |
| 34 | Anacardiaceae | Toxicodendron | radicans | Eastern poison ivy | Native | Forb/Herb/Shrub/Subshrub/Vine | Perennial |
| 35 | Ericaceae | Vaccinium | angustifolium | Lowbush blueberry | Native | Shrub/Subshrub | Perennial |
| 36 | Vitaceae | Vitis | vinifera | Wine grape | Introduced | Shrub/Vine | Perennial |
| 37 | Rutaceae | Zanthoxylum | simulans | Chinese-pepper | Introduced | Shrub/Tree | Perennial |
| Plant Trait Group | Full Plant Trait Name 1 | Total Number of Records per Dataset | Plant Origin 2 | Number of Plant Species per Dataset | Mean ± SE | Units |
|---|---|---|---|---|---|---|
| Leaf characteristics | Leaf area (in case of compound leaves: leaf, petiole excluded) | 2537 | I * | 1 | 778.56 ± 161.45 | cm2 |
| N | 12 | 52.63 ± 0.51 | cm2 | |||
| Leaf area (in case of compound leaves: leaf, petiole included) | 2740 | I | 5 | 155.73 ± 21.11 | cm2 | |
| N | 20 | 53.63 ± 0.63 | cm2 | |||
| Leaf dry mass (single leaf) | 5035 | I | 6 | 1.03 ± 0.21 | g | |
| N | 18 | 0.16 ± 0 | g | |||
| Leaf fresh mass | 4716 | I | 3 | 3.07 ± 0.59 | g | |
| N | 12 | 0.45 ± 0.01 | g | |||
| Leaf chlorophyll content per leaf area | 313 | I * | 1 | 26.32 ± 1.09 | CCM | |
| N | 5 | 13.88 ± 0.14 | CCM | |||
| Leaf length | 160 | I | 2 | 0.92 ± 0.22 | cm | |
| N | 7 | 1.05 ± 0.04 | cm | |||
| Leaf nitrogen (N) content per leaf | 21 | I | 4 | 8.35 ± 2.43 | g m−2 | |
| N | 8 | 11.95 ± 3.06 | g m−2 | |||
| Leaf nitrogen (N) content per leaf dry mass | 3320 | I | 17 | 0.03 ± 0 | g/g | |
| N | 19 | 0.02 ± 0 | g/g | |||
| Leaf water content per leaf dry mass (not saturated) | 3320 | I | 12 | 25.48 ± 0.32 | g/g | |
| N | 11 | 22.6 ± 0.12 | g/g | |||
| Plant architecture and lifespan | Crown (canopy) length: diameter along the longest axis | 146 | I * | 1 | 0.98 ± 0.05 | m |
| N | 4 | 0.07 ± 0 | m | |||
| Crown (canopy) height | 117 | I | 4 | 3.5 ± 0.51 | m | |
| N | 1 | 0.33 ± 0 | m | |||
| Crown (canopy) width | 146 | I * | 1 | 0.98 ± 0.06 | m | |
| N | 4 | 0.05 ± 0 | m | |||
| Plant lifespan (longevity) max | 21 | I | 4 | 185.83 ± 51.31 | years | |
| N | 7 | 324.56 ± 96.3 | years | |||
| Plant lifespan (longevity) mean | 36 | I | 3 | 310 ± 55.54 | years | |
| N | 11 | 240.65 ± 36.76 | years | |||
| Plant lifespan (longevity) min | 16 | I | 4 | 174 ± 50.69 | years | |
| N | 4 | 157.5 ± 47.08 | years | |||
| Plant height vegetative max | 442 | I | 17 | 22.85 ± 0.74 | m | |
| N | 19 | 25.46 ± 0.65 | m | |||
| Stem diameter | 2609 | I | 9 | 0.25 ± 0.02 | m | |
| N | 16 | 0.21 ± 0 | m | |||
| Species habitat characterization/Plant requirements | Precipitation max | 25 | I | 6 | 54.17 ± 2.01 | in/ft |
| N | 19 | 57.5 ± 2.5 | ||||
| Precipitation min | 25 | I | 6 | 31.33 ± 0.42 | in/ft | |
| N | 19 | 28.5 ± 2.03 | ||||
| Temperature: species environmental indicator value according to Ellenberg | 20 | I | 8 | 5.5 ± 0.54 | EV 3 | |
| N | 3 | 5.8 ± 0.49 | EV | |||
| soil pH max | 25 | I | 6 | 7.28 ± 0.1 | pH value | |
| N | 19 | 6.98 ± 0.25 | ||||
| soil pH” min | 25 | I | 6 | 5.13 ± 0.14 | pH value | |
| N | 19 | 4.98 ± 0.17 | ||||
| Light: species environmental indicator value according to Ellenberg | 23 | I | 8 | 5 ± 0.53 | EV | |
| N | 4 | 4.29 ± 0.71 | EV | |||
| Atmospheric CO2 concentration | 193 | I | 3 | 435.28 ± 32.28 | ppm | |
| N | 10 | 379.18 ± 6.54 | ppm | |||
| Elevation, m | 627 | I | 8 | 238.32 ± 9.77 | m | |
| N | 14 | 271.64 ± 8.82 | m |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avanesyan, A.; McPherson, C.; Lamp, W.O. Analysis of Plant Trait Data of Host Plants of Lycorma delicatula in the US Suggests Evidence for Ecological Fitting. Forests 2022, 13, 2017. https://doi.org/10.3390/f13122017
Avanesyan A, McPherson C, Lamp WO. Analysis of Plant Trait Data of Host Plants of Lycorma delicatula in the US Suggests Evidence for Ecological Fitting. Forests. 2022; 13(12):2017. https://doi.org/10.3390/f13122017
Chicago/Turabian StyleAvanesyan, Alina, Cameron McPherson, and William O. Lamp. 2022. "Analysis of Plant Trait Data of Host Plants of Lycorma delicatula in the US Suggests Evidence for Ecological Fitting" Forests 13, no. 12: 2017. https://doi.org/10.3390/f13122017