Unimolecular Micelles from Randomly Grafted Arborescent Copolymers with Different Core Branching Densities: Encapsulation of Doxorubicin and In Vitro Release Study
Abstract
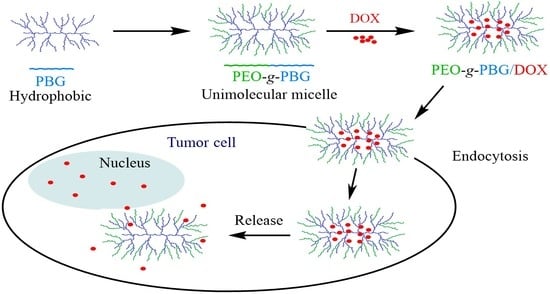
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Characterization
2.3. Synthetic Procedures
2.3.1. Synthesis of G0 Arborescent PBG
2.3.2. Synthesis of Arborescent Copolymers
2.4. Loading of DOX in the Unimolecular Micelles
2.5. In Vitro Release of DOX
3. Results and Discussion
3.1. Synthesis of Linear PBG Substrates
3.2. Synthesis of G0PBG with Different Branching Densities
3.3. Synthesis of Randomly Grafted Arborescent Copolymers
Properties of the Arborescent Copolymer Micelles
3.4. Drug Loading and Micelle Characterization
3.5. In Vitro Drug Release Kinetics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perin, F.; Motta, A.; Maniglio, D. Amphiphilic copolymers in biomedical applications: Synthesis routes and property control. Mater. Sci. Eng. C 2021, 123, 111952. [Google Scholar] [CrossRef] [PubMed]
- Kuperkar, K.; Patel, D.; Atanase, L.I.; Bahadur, P. Amphiphilic Block Copolymers: Their Structures, and Self-Assembly to Polymeric Micelles and Polymersomes as Drug Delivery Vehicles. Polymers 2022, 14, 4702. [Google Scholar] [CrossRef]
- Avramović, N.; Mandić, B.; Savić-Radojević, A.; Simić, T. Polymeric Nanocarriers of Drug Delivery Systems in Cancer Therapy. Pharmaceutics 2020, 12, 298. [Google Scholar] [CrossRef] [Green Version]
- Ghezzi, M.; Pescina, S.; Padula, C.; Santi, P.; Del Favero, E.; Cantù, L.; Nicoli, S. Polymeric micelles in drug delivery: An insight of the techniques for their characterization and assessment in biorelevant conditions. J. Control. Release 2021, 332, 312–336. [Google Scholar] [CrossRef]
- Alsehli, M. Polymeric nanocarriers as stimuli-responsive systems for targeted tumor (cancer) therapy: Recent advances in drug delivery. Saudi Pharm. J. 2020, 28, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Chytil, P.; Koziolova, E.; Janouskova, O.; Kostka, L.; Ulbrich, K.; Etrych, T. Synthesis and Properties of Star HPMA Copolymer Nanocarriers Synthesised by RAFT Polymerisation Designed for Selective Anticancer Drug Delivery and Imaging. Macromol. Biosci. 2015, 15, 839–850. [Google Scholar] [CrossRef]
- Gong, J.; Chen, M.; Zheng, Y.; Wang, S.; Wang, Y. Polymeric micelles drug delivery system in oncology. J. Control. Release 2012, 159, 312–323. [Google Scholar] [CrossRef]
- Khandare, J.; Calderon, M.; Dagia, N.M.; Haag, R. Multifunctional dendritic polymers in nanomedicine: Opportunities and challenges. Chem. Soc. Rev. 2012, 41, 2824–2848. [Google Scholar] [CrossRef] [Green Version]
- Kurniasih, I.N.; Keilitz, J.; Haag, R. Dendritic nanocarriers based on hyperbranched polymers. Chem. Soc. Rev. 2015, 44, 4145–4164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukowiak, M.C.; Thota, B.N.S.; Haag, R. Dendritic core–shell systems as soft drug delivery nanocarriers. Biotechnol. Adv. 2015, 33, 1327–1341. [Google Scholar] [CrossRef]
- Alsehli, M.; Gauthier, M. Dendritic Polymer Micelles for Drug Delivery. In Bioinspired Materials Science and Engineering; John Wiley: Hoboken, NJ, USA, 2018; pp. 311–335. [Google Scholar] [CrossRef]
- Paolino, M.; Ennen, F.; Lamponi, S.; Cernescu, M.; Voit, B.; Cappelli, A.; Appelhans, D.; Komber, H. Cyclodextrin-Adamantane Host–Guest Interactions on the Surface of Biocompatible Adamantyl-Modified Glycodendrimers. Macromolecules 2013, 46, 3215–3227. [Google Scholar] [CrossRef]
- Kannan, R.M.; Nance, E.; Kannan, S.; Tomalia, D.A. Emerging concepts in dendrimer-based nanomedicine: From design principles to clinical applications. J. Intern. Med. 2014, 276, 579–617. [Google Scholar] [CrossRef]
- Wu, W.; Wang, W.; Li, J. Star polymers: Advances in biomedical applications. Prog. Polym. Sci. 2015, 46, 55–85. [Google Scholar] [CrossRef]
- Sowinska, M.; Urbanczyk-Lipkowska, Z. Advances in the chemistry of dendrimers. New J. Chem. 2014, 38, 2168–2203. [Google Scholar] [CrossRef]
- Wu, L.-p.; Ficker, M.; Christensen, J.B.; Trohopoulos, P.N.; Moghimi, S.M. Dendrimers in Medicine: Therapeutic Concepts and Pharmaceutical Challenges. Bioconjugate. Chem. 2015, 26, 1198–1211. [Google Scholar] [CrossRef] [PubMed]
- Kurtoglu, Y.E.; Mishra, M.K.; Kannan, S.; Kannan, R.M. Drug release characteristics of PAMAM dendrimer–drug conjugates with different linkers. Int. J. Pharm. 2010, 384, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Tomalia, D.A.; Christensen, J.B.; Boas, U. Dendrimers, Dendrons, and Dendritic Polymers; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Sun, T.; Zhang, Y.S.; Pang, B.; Hyun, D.C.; Yang, M.; Xia, Y. Engineered Nanoparticles for Drug Delivery in Cancer Therapy. Angew. Chem. Int. Ed. 2014, 53, 12320–12364. [Google Scholar] [CrossRef]
- Svenson, S. The dendrimer paradox—High medical expectations but poor clinical translation. Chem. Soc. Rev. 2015, 44, 4131–4144. [Google Scholar] [CrossRef]
- Gauthier, M.; Möller, M. Uniform highly branched polymers by anionic grafting: Arborescent graft polymers. Macromolecules 1991, 24, 4548–4553. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Hedstrand, D.M.; Ferritto, M.S. Comb-burst dendrimer topology: New macromolecular architecture derived from dendritic grafting. Macromolecules 1991, 24, 1435–1438. [Google Scholar] [CrossRef]
- Gauthier, M. Arborescent polymers and other dendrigraft polymers: A journey into structural diversity. J. Polym. Sci. Part A Polym. Chem. 2007, 45, 3803–3810. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Njikang, G.; Gauthier, M.; Li, J. Arborescent polystyrene-graft-poly(2-vinylpyridine) copolymers as unimolecular micelles: Solubilization studies. Polymer 2008, 49, 1276–1284. [Google Scholar] [CrossRef]
- Njikang, G.N.; Gauthier, M.; Li, J. Sustained release properties of arborescent polystyrene-graft-poly(2-vinylpyridine) copolymers. Polymer 2008, 49, 5474–5481. [Google Scholar] [CrossRef]
- Whitton, G.; Gauthier, M. Arborescent Polypeptides from γ-Benzyl L-Glutamic Acid. J. Polym. Sci. A Polym. Chem. 2013, 51, 5270–5279. [Google Scholar] [CrossRef]
- Whitton, G.; Gauthier, M. Arborescent micelles: Dendritic poly(γ-benzyl l-glutamate) cores grafted with hydrophilic chain segments. J. Polym. Sci. Part A Polym. Chem. 2016, 54, 1197–1209. [Google Scholar] [CrossRef]
- Ji, S.; Hoye, T.R.; Macosko, C.W. Primary Amine (−NH2) Quantification in Polymers: Functionality by 19F NMR Spectroscopy. Macromolecules 2005, 38, 4679–4686. [Google Scholar] [CrossRef]
- Poché, D.S.; Moore, M.J.; Bowles, J.L. An Unconventional Method for Purifying the N-carboxyanhydride Derivatives of γ-alkyl-L-glutamates. Synth. Commun. 1999, 29, 843–854. [Google Scholar] [CrossRef]
- Alsehli, M.; Gauthier, M. Arborescent Polypeptides for Sustained Drug Delivery. MRS Online Proc. Libr. (OPL) 2016, 1819, imrc2015s2014d-o2015. [Google Scholar] [CrossRef]
- Geng, Y.; Dalhaimer, P.; Cai, S.; Tsai, R.; Tewari, M.; Minko, T.; Discher, D.E. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2007, 2, 249–255. [Google Scholar] [CrossRef]
- Cai, S.; Vijayan, K.; Cheng, D.; Lima, E.M.; Discher, D.E. Micelles of different morphologies—Advantages of worm-like filomicelles of PEO-PCL in paclitaxel delivery. Pharm. Res. 2007, 24, 2099–2109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malm, A.V.; Corbett, J.C.W. Improved dynamic light scattering using an adaptive and statistically driven time resolved treatment of correlation data. Sci. Rep. 2019, 9, 13519. [Google Scholar] [CrossRef] [Green Version]
- Chow, E.K.; Ho, D. Cancer nanomedicine: From drug delivery to imaging. Sci. Transl. Med. 2013, 5, 216rv214. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, S.; Liu, H.; Yang, C.; Kang, Y.; Wang, M. Unimolecular micelles of amphiphilic cyclodextrin-core star-like block copolymers for anticancer drug delivery. Chem. Commun. 2015, 51, 15768–15771. [Google Scholar] [CrossRef]
- Pouton, C.W.; Wagstaff, K.M.; Roth, D.M.; Moseley, G.W.; Jans, D.A. Targeted delivery to the nucleus. Adv. Drug Deliv. Rev. 2007, 59, 698–717. [Google Scholar] [CrossRef] [PubMed]
- Prabaharan, M.; Grailer, J.J.; Pilla, S.; Steeber, D.A.; Gong, S. Gold nanoparticles with a monolayer of doxorubicin-conjugated amphiphilic block copolymer for tumor-targeted drug delivery. Biomaterials 2009, 30, 6065–6075. [Google Scholar] [CrossRef] [PubMed]
- Bareford, L.M.; Swaan, P.W. Endocytic mechanisms for targeted drug delivery. Adv. Drug Deliv. Rev. 2007, 59, 748–758. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.-M.; Chen, Y.; Hou, X.-F.; Wu, X.; Gu, B.-H.; Liu, Y. Sulfonato-β-cyclodextrin mediated supramolecular nanoparticle for controlled release of berberine. ACS Appl. Mater. Interfaces 2018, 10, 24987–24992. [Google Scholar] [CrossRef]
- Normant, H.; Angelo, B. Sodation en milieu tétrahydrofuranne par le sodium en présence de naphthalène. Bulletin De La Société Chimique De France 1960, 354–359. [Google Scholar]










| Sample | Deprotection Level of Substrate (%) | Molar Ratio CO2H:NH2 | Mn(g/mol) | Ð | Gy (%) | Ce (%) | fn | bd |
|---|---|---|---|---|---|---|---|---|
| G0PBG15 | 100 | 1.1:1 | 65,400 | 1.04 | 76 | 80 | 12 | 0.80 |
| G0PBG29 | 31 | 1.7:1 | 35,300 | 1.08 | 87 | 54 | 5.4 | 0.19 |
| G0PBG65 | 31 | 1.7:1 | 51,000 | 1.06 | 89 | 36 | 7.1 | 0.11 |
| Copolymer | PBG Substrate | Graft Copolymer | |||||
|---|---|---|---|---|---|---|---|
| Mn (g/mol) a | %-CO2H b | Gy c | Mn (g/mol) a | Mw/Mn a | fnd | %-PEO e | |
| G1PBG15-g-PEO | 240,000 | 31 | 27 | 1.2 × 106 | 1.07 | 96 | 80 |
| G1PBG29-g-PEO | 200,000 | 31 | 27 | 1.0 × 106 | 1.09 | 80 | 80 |
| G1PBG65-g-PEO | 260,000 | 33 | 29 | 1.5 × 106 | 1.08 | 124 | 82 |
| G2PBG15-g-PEO | 1.2 × 106 | 31 | 13 | 3.4 × 106 | 1.08 | 220 | 65 |
| G2PBG29-g-PEO | 1.0 × 106 | 32 | 15 | 3.2 × 106 | 1.09 | 220 | 69 |
| G2PBG65-g-PEO | 1.4 × 106 | 30 | 13 | 3.9 × 106 | 1.06 | 250 | 64 |
| Copolymer | G2PBG-g-PEO | G2PBG-g-PEO/DOX | ||||
|---|---|---|---|---|---|---|
| Size (nm) | PDI | Size (nm) | PDI | DLC (wt%) | DLE (%) | |
| G1PBG15-g-PEO/DOX | 16 | 0.41 | 17 | 0.38 | 8.3 ± 0.3 | 50 ± 2 |
| G1PBG29-g-PEO/DOX | 17 | 0.51 | 17 | 0.54 | 7.1 ± 0.2 | 42 ± 3 |
| G1PBG65-g-PEO/DOX | 19 | 0.43 | 20 | 0.44 | 7.4 ± 0.2 | 44 ± 4 |
| G2PBG15-g-PEO/DOX | 35 | 0.28 | 37 | 0.49 | 10.8 ± 0.2 | 65 ± 3 |
| G2PBG29-g-PEO/DOX | 34 | 0.26 | 35 | 0.51 | 10.1 ± 0.2 | 60 ± 3 |
| G2PBG65-g-PEO/DOX | 39 | 0.17 | 41 | 0.47 | 10.3 ± 0.3 | 62 ± 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsehli, M.; Gauthier, M. Unimolecular Micelles from Randomly Grafted Arborescent Copolymers with Different Core Branching Densities: Encapsulation of Doxorubicin and In Vitro Release Study. Materials 2023, 16, 2461. https://doi.org/10.3390/ma16062461
Alsehli M, Gauthier M. Unimolecular Micelles from Randomly Grafted Arborescent Copolymers with Different Core Branching Densities: Encapsulation of Doxorubicin and In Vitro Release Study. Materials. 2023; 16(6):2461. https://doi.org/10.3390/ma16062461
Chicago/Turabian StyleAlsehli, Mosa, and Mario Gauthier. 2023. "Unimolecular Micelles from Randomly Grafted Arborescent Copolymers with Different Core Branching Densities: Encapsulation of Doxorubicin and In Vitro Release Study" Materials 16, no. 6: 2461. https://doi.org/10.3390/ma16062461