The UV Dose Used for Disinfection of Drinking Water in Sweden Inadequately Inactivates Enteric Virus with Double-Stranded Genomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Host Cells and Virus Stocks
2.2. Viral Infectivity Titers
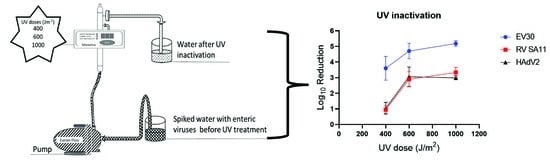
2.3. UV Light Inactivation
2.4. Detection of Viral Nucleic Acid by qPCR
2.5. Statistical Analysis
3. Results
3.1. Viral Inactivation
3.2. Confirmation That the Correct Virus Was Used in the Experiment
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Primers | Primer Sequences | Reference |
|---|---|---|
| HAdV2-forward | GCCACGGTGGGGTTTCTAAACTT | [25] |
| HAdV2-reverse | GCCCCAGTGGTCTTACATGCACATC | [25] |
| HAdV2-probe | [FAM] TGCACCAGACCCGGGCTCAGGTACTCCGA [MGBEQ] | [25] |
| EV30-forward | TCCTCCGGCCCCTGAATGCG | [28] |
| EV30-reverse1 | ACCGGATGGCCAATCCAA | [28] |
| EV30-reverse2 | ATGTCACCATAAGCAGCCA | [28] |
| EV30-probe | [FAM] CGGAACCGACTACTTTGGGTGIC [MGBEQ] | [28] |
| RV SA11-forward | ACCATCTWCACRTRACCCTCTATGAGA | [47] |
| RV SA11-reverse | GGTCACATAACGCCCCTATAGC | [47] |
| RV SA11-probe | [FAM] AGTTAAAAGCTAACACTGTCAAA [MGBEQ] | [47] |
| Dilution | EV30 | RV SA11 | HAdV2 | |||
|---|---|---|---|---|---|---|
| Cell Culture a | rt-qPCR b | Cell Culture a | rt-qPCR b | Cell Culture a | rt-qPCR b | |
| 100 | + | + | + | + | + | + |
| 10−1 | + | + | + | + | + | + |
| 10−2 | + | + | + | + | + | + |
| 10−3 | + | + | + | − | + | + |
| 10−4 | + | − | + | − | + | − |
| 10−5 | + | − | + | − | + | − |
| 10−6 | + | − | − | − | − | − |
| 10−7 | − | − | − | − | − | − |
References
- Chitambar, S.; Gopalkrishna, V.; Chhabra, P.; Patil, P.; Verma, H.; Lahon, A.; Arora, R.; Tatte, V.; Ranshing, S.; Dhale, G.; et al. Diversity in the enteric viruses detected in outbreaks of gastroenteritis from Mumbai, Western India. Int. J. Environ. Res. Public Health 2012, 9, 895–915. [Google Scholar] [CrossRef] [PubMed]
- EFSA. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 2015, 13, 4329. [Google Scholar]
- Carol, M.; Guadalupe-Fernández, V.; Rius, C.; Soldevila, N.; Razquin, E.; Guix, S.; Dominguez, A.; on behalf of the Working Group for the Study of Outbreaks of Acute Gastroenteritis in Catalonia. A Waterborne Gastroenteritis Outbreak Caused by a GII Norovirus in a Holiday Camp in Catalonia (Spain), 2017. Viruses 2021, 13, 1792. [Google Scholar] [CrossRef] [PubMed]
- Brunkard, J.M.; Ailes, E.; A Roberts, V.; Hill, V.; Hilborn, E.D.; Craun, G.F.; Rajasingham, A.; Kahler, A.; Garrison, L.; Hicks, L.; et al. Surveillance for waterborne disease outbreaks associated with drinking water—United States, 2007–2008. MMWR Surveill. Summ. 2011, 60, 38–68. [Google Scholar] [PubMed]
- Verheyen, J.; Timmen-Wego, M.; Laudien, R.; Boussaad, I.; Sen, S.; Koc, A.; Uesbeck, A.; Mazou, F.; Pfister, H. Detection of adenoviruses and rotaviruses in drinking water sources used in rural areas of Benin, West Africa. Appl. Environ. Microbiol. 2009, 75, 2798–2801. [Google Scholar] [CrossRef] [Green Version]
- Ødegaard, E.H.; Østerhus, S.W.; Pott, B.M. Microbial Barrier Analysis (MBA—A Guideline); Norwegian Water BA: Hamar, Norway, 2014; p. 74. [Google Scholar]
- Hijnen, W.A.; Beerendonk, E.F.; Medema, G.J. Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: A review. Water Res. 2006, 40, 3–22. [Google Scholar] [CrossRef]
- Haider, T.; Sommer, R.; Knasmüller, S.; Eckl, P.; Pribil, W.; Cabaj, A.; Kundi, M. Genotoxic response of Austrian groundwater samples treated under standardized UV (254 nm)—Disinfection conditions in a combination of three different bioassays. Water Res. 2002, 36, 25–32. [Google Scholar] [CrossRef]
- Douki, T. The variety of UV-induced pyrimidine dimeric photoproducts in DNA as shown by chromatographic quantification methods. Photochem. Photobiol. Sci. 2013, 12, 1286–1302. [Google Scholar] [CrossRef]
- Ko, G.; Cromeans, T.L.; Sobsey, M.D. UV inactivation of adenovirus type 41 measured by cell culture mRNA RT-PCR. Water Res. 2005, 39, 3643–3649. [Google Scholar] [CrossRef]
- Araud, E.; Fuzawa, M.; Shisler, J.L.; Li, J.; Nguyen, T.H. UV Inactivation of Rotavirus and Tulane Virus Targets Different Components of the Virions. Appl. Environ. Microbiol. 2020, 86, 4. [Google Scholar] [CrossRef]
- Simonet, J.; Gantzer, C. Inactivation of poliovirus 1 and F-specific RNA phages and degradation of their genomes by UV irradiation at 254 nanometers. Appl. Environ. Microbiol. 2006, 72, 7671–7677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, G.W.; Linden, K.G.; Sobsey, M.D. Inactivation of murine norovirus, feline calicivirus and echovirus 12 as surrogates for human norovirus (NoV) and coliphage (F+) MS2 by ultraviolet light (254 nm) and the effect of cell association on UV inactivation. Lett. Appl. Microbiol. 2011, 52, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Z.; Ye, Y.; Chang, P.H.; Thirunarayanan, D.; Wigginton, K.R. Nucleic Acid Photolysis by UV(254) and the Impact of Virus Encapsidation. Environ. Sci. Technol. 2018, 52, 10408–10415. [Google Scholar] [CrossRef] [PubMed]
- Calgua, B.; Carratalà, A.; Guerrero-Latorre, L.; Corrêa, A.d.A.; Kohn, T.; Sommer, R.; Girones, R. UVC Inactivation of dsDNA and ssRNA Viruses in Water: UV Fluences and a qPCR-Based Approach to Evaluate Decay on Viral Infectivity. Food Environ. Virol. 2014, 6, 260–268. [Google Scholar] [CrossRef]
- Li, D.; Gu, A.Z.; He, M.; Shi, H.-C.; Yang, W. UV inactivation and resistance of rotavirus evaluated by integrated cell culture and real-time RT-PCR assay. Water Res. 2009, 43, 3261–3269. [Google Scholar] [CrossRef]
- Pang, X.; Qiu, Y.; Gao, T.; Zurawell, R.; Neumann, N.F.; Craik, S.; Lee, B.E. Prevalence, levels and seasonal variations of human enteric viruses in six major rivers in Alberta, Canada. Water Res. 2019, 153, 349–356. [Google Scholar] [CrossRef]
- Borchardt, M.A.; Haas, N.L.; Hunt, R.J. Vulnerability of drinking-water wells in La Crosse, Wisconsin, to enteric-virus contamination from surface water contributions. Appl. Environ. Microbiol. 2004, 70, 5937–5946. [Google Scholar] [CrossRef] [Green Version]
- Bortagaray, V.; Girardi, V.; Pou, S.; Lizasoain, A.; Tort, L.F.L.; Spilki, F.R.; Colina, R.; Victoria, M. Detection, Quantification, and Microbial Risk Assessment of Group A Rotavirus in Rivers from Uruguay. Food Environ. Virol. 2020, 12, 89–98. [Google Scholar] [CrossRef]
- Miura, T.; Gima, A.; Akiba, M. Detection of Norovirus and Rotavirus Present in Suspended and Dissolved Forms in Drinking Water Sources. Food Environ. Virol. 2019, 11, 9–19. [Google Scholar] [CrossRef]
- Stokdyk, J.P.; Firnstahl, A.D.; Walsh, J.F.; Spencer, S.K.; de Lambert, J.R.; Anderson, A.C.; Rezania, L.-I.W.; Kieke, B.A.; Borchardt, M.A. Viral, bacterial, and protozoan pathogens and fecal markers in wells supplying groundwater to public water systems in Minnesota, USA. Water Res. 2020, 178, 115814. [Google Scholar] [CrossRef]
- Pang, X.; Gao, T.; Qiu, Y.; Caffrey, N.; Popadynetz, J.; Younger, J.; Lee, B.E.; Neumann, N.; Checkley, S. The prevalence and levels of enteric viruses in groundwater of private wells in rural Alberta, Canada. Water Res. 2021, 202, 117425. [Google Scholar] [CrossRef] [PubMed]
- Gratacap-Cavallier, B.; Genoulaz, O.; Brengel-Pesce, K.; Soule, H.; Innocenti-Francillard, P.; Bost, M.; Gofti, L.; Zmirou, D.; Seigneurin, J.M. Detection of human and animal rotavirus sequences in drinking water. Appl. Environ. Microbiol. 2000, 66, 2690–2692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashid, M.; Khan, M.N.; Jalbani, N. Detection of Human Adenovirus, Rotavirus, and Enterovirus in Tap Water and Their Association with the Overall Quality of Water in Karachi, Pakistan. Food Environ. Virol. 2021, 13, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sikora, P.; Rutgersson, C.; Lindh, M.; Brodin, T.; Björlenius, B.; Larsson, D.J.; Norder, H. Differential removal of human pathogenic viruses from sewage by conventional and ozone treatments. Int. J. Hyg. Environ. Health 2018, 221, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- ATCC. Is It Possible to Determine from the Tcid[50] How Many Plaque Forming Units to Expect? 2019. Available online: https://www.atcc.org/support/technical-support/faqs/converting-tcid-50-to-plaque-forming-units-pfu (accessed on 7 December 2021).
- Wang, H.; Neyvaldt, J.; Enache, L.; Sikora, P.; Mattsson, A.; Johansson, A.; Lindh, M.; Bergstedt, O.; Norder, H. Variations among Viruses in Influent Water and Effluent Water at a Wastewater Plant over One Year as Assessed by Quantitative PCR and Metagenomics. Appl. Environ. Microbiol. 2020, 86, e02073-20. [Google Scholar] [CrossRef]
- U.S.EPA. Ultraviolet Disinfection Guidance Manual for the Final Long Term 2 Enhanced Surface Water Treatment Rule; Office of Water: Washington, DC, USA, 2006; Volume 20460, p. 53.
- DVGW. DVGW Work Sheet W 294: UV Disinfection Devices for Drinking Water Supplies—Requirements and Testing. DVGW. 1997. Available online: https://infostore.saiglobal.com/en-us/standards/dvgw-w-294-1997-491296_saig_dvgw_dvgw_1100317/ (accessed on 10 December 2021).
- ÖNORM. Plants for Disinfection of Water Using Ultravioloet Radiation—Requirements and Testing—Part 1: Low Pressure Mercury Lamp Plants; M 5873 Revised Version; Austrian Standards International: Wien, Austria, 2000. [Google Scholar]
- Eriksson, U. P117 Advice and Guidelines on UV Light for Water Treatment; Svenskt Vatten AB: Stockholm, Sweden, 2021; p. 38. [Google Scholar]
- Gerba, C.P.; Gramos, D.M.; Nwachuku, N. Comparative inactivation of enteroviruses and adenovirus 2 by UV light. Appl. Environ. Microbiol. 2002, 68, 5167–5169. [Google Scholar] [CrossRef] [Green Version]
- Ballester, N.A.; Malley, J.P. Sequential disinfection of adenovirus type 2 with UV-chlorine-chloramine. J. Am. Water Work. Assoc. 2004, 96, 97–103. [Google Scholar] [CrossRef]
- Eischeid, A.C.; Meyer, J.N.; Linden, K.G. UV disinfection of adenoviruses: Molecular indications of DNA damage efficiency. Appl. Environ. Microbiol. 2009, 75, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Baxter, C.S.; Hofmann, R.; Templeton, M.R.; Brown, M.; Andrews, R.C. Inactivation of adenovirus Types 2, 5, and 41 in drinking water by UV light, free chlorine, and monochloramine. J. Environ. Eng. 2007, 133, 95–103. [Google Scholar] [CrossRef] [Green Version]
- Shin, G.A.; Lee, J.K.; Linden, K.G. Enhanced effectiveness of medium-pressure ultraviolet lamps on human adenovirus 2 and its possible mechanism. Water Sci. Technol. 2009, 60, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, R.A.; Bounty, S.; Linden, K.G. Long-range quantitative PCR for determining inactivation of adenovirus 2 by ultraviolet light. J. Appl. Microbiol. 2013, 114, 1854–1865. [Google Scholar] [CrossRef] [PubMed]
- Bounty, S.; Rodriguez, R.A.; Linden, K.G. Inactivation of adenovirus using low-dose UV/H2O2 advanced oxidation. Water Res. 2012, 46, 6273–6278. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C.; Ossoff, S.F.; Lobe, D.C.; Dorfman, M.H.; Dumais, C.M.; Qualls, R.G.; Johnson, J.D. UV inactivation of pathogenic and indicator microorganisms. Appl. Environ. Microbiol. 1985, 49, 1361–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caballero, S.; Abad, F.X.; Loisy, F.; Le Guyader, F.S.; Cohen, J.; Pintó, R.M.; Bosch, A. Rotavirus virus-like particles as surrogates in environmental persistence and inactivation studies. Appl. Environ. Microbiol. 2004, 70, 3904–3909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, S.E.; Rodriguez, R.A.; Hawkins, M.A.; Hargy, T.M.; Larason, T.C.; G, K. Linden Comparison of UV-Induced Inactivation and RNA Damage in MS2 Phage across the Germicidal UV Spectrum. Appl. Environ. Microbiol. 2015, 82, 1468–1474. [Google Scholar] [CrossRef] [Green Version]
- WHO. Guidelines for Drinking-Water Quality; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Guo, H.; Chu, X.; Hu, J. Effect of host cells on low- and medium-pressure UV inactivation of adenoviruses. Appl. Environ. Microbiol. 2010, 76, 7068–7075. [Google Scholar] [CrossRef] [Green Version]
- Thurman, R.B.; Gerba, C.P. Molecular mechanisms of viral inactivation by water disinfectants. Adv. Appl. Microbiol. 1988, 33, 75–105. [Google Scholar]
- Oh, C.; Sun, P.P.; Araud, E.M.; Nguyen, T.H. Mechanism and efficacy of virus inactivation by a microplasma UV lamp generating monochromatic UV irradiation at 222 nm. Water Res. 2020, 186, 116386. [Google Scholar] [CrossRef]
- Zeng, S.-Q.; Halkosalo, A.; Salminen, M.; Szakal, E.; Puustinen, L.; Vesikari, T. One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis. J. Virol. Methods 2008, 153, 238–240. [Google Scholar] [CrossRef]
| Virus | 400 J/m2 | 600 J/m2 | 1000 J/m2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Flow Rate (L/s) | Detention Time (s) | UV Dose (J/m2) | Flow Rate (L/s) | Detention Time (s) | UV Dose (J/m2) | Flow Rate (L/s) | Detention Time (s) | UV Dose (J/m2) | ||||
| Max | Min | Max | Min | Max | Min | |||||||
| EV30 | 0.17 | 11.28 | 448 | 403 | 0.14 | 12.75 | 506 | 456 | 0.08 | 23.50 | 1011 | 910 |
| 0.18 | 10.79 | 429 | 386 | 0.12 | 15.11 | 600 | 540 | 0.08 | 23.39 | 1006 | 906 | |
| RV SA11 | 0.20 | 9.94 | 395 | 356 | 0.13 | 14.19 | 564 | 507 | 0.08 | 22.60 | 972 | 875 |
| 0.20 | 9.94 | 395 | 356 | 0.12 | 14.57 | 579 | 521 | 0.08 | 23.81 | 1024 | 922 | |
| 0.20 | 9.94 | 395 | 356 | 0.12 | 14.57 | 579 | 521 | 0.08 | 23.81 | 1024 | 922 | |
| HAdV2 | 0.19 | 10.10 | 401 | 361 | 0.12 | 14.84 | 590 | 531 | 0.08 | 23.41 | 1007 | 906 |
| 0.20 | 9.94 | 395 | 356 | 0.12 | 14.84 | 590 | 531 | 0.08 | 23.34 | 1004 | 904 | |
| 0.19 | 10.38 | 412 | 371 | 0.12 | 14.64 | 582 | 524 | 0.08 | 23.50 | 1011 | 910 | |
| 0.19 | 10.38 | 412 | 371 | 0.13 | 14.08 | 560 | 504 | 0.08 | 23.50 | 1011 | 910 | |
| Virus | UV Dose Range (J/m2) | Initial Viral Concentration | Log10 Reduction | ANOVA | |||
|---|---|---|---|---|---|---|---|
| Min | Max | Mean (TCID50/mL) | Mean | Min | Max | (p-Value) | |
| EV30 | 386 | 448 | 9.05 × 104 | 3.60 | 2.84 | 4.36 | |
| 506 | 600 | 8.70 × 104 | 4.70 | 4.20 | 5.20 | ||
| 906 | 1011 | 1.39 × 105 | 5.18 | 5.00 | 5.36 | 0.249 | |
| RV SA11 | 356 | 395 | 9.29 × 105 | 0.95 | 0.50 | 1.34 | |
| 507 | 579 | 5.99 × 105 | 2.89 | 2.00 | 3.67 | ||
| 875 | 1024 | 7.70 × 105 | 3.33 | 3.00 | 4.00 | 0.008 | |
| HAdV2 | 356 | 412 | 1.66 × 103 | 1.04 | 0.16 | 2.00 | |
| 504 | 590 | 1.36 × 104 | 3.06 | 2.00 | 4.70 | ||
| 904 | 1011 | 9.17 × 102 | 2.99 | 2.70 | 3.20 | 0.013 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saguti, F.; Churqui, M.P.; Kjellberg, I.; Wang, H.; Ottoson, J.; Paul, C.; Bergstedt, O.; Norder, H.; Nyström, K. The UV Dose Used for Disinfection of Drinking Water in Sweden Inadequately Inactivates Enteric Virus with Double-Stranded Genomes. Int. J. Environ. Res. Public Health 2022, 19, 8669. https://doi.org/10.3390/ijerph19148669
Saguti F, Churqui MP, Kjellberg I, Wang H, Ottoson J, Paul C, Bergstedt O, Norder H, Nyström K. The UV Dose Used for Disinfection of Drinking Water in Sweden Inadequately Inactivates Enteric Virus with Double-Stranded Genomes. International Journal of Environmental Research and Public Health. 2022; 19(14):8669. https://doi.org/10.3390/ijerph19148669
Chicago/Turabian StyleSaguti, Fredy, Marianela Patzi Churqui, Inger Kjellberg, Hao Wang, Jakob Ottoson, Catherine Paul, Olof Bergstedt, Heléne Norder, and Kristina Nyström. 2022. "The UV Dose Used for Disinfection of Drinking Water in Sweden Inadequately Inactivates Enteric Virus with Double-Stranded Genomes" International Journal of Environmental Research and Public Health 19, no. 14: 8669. https://doi.org/10.3390/ijerph19148669