Fast and Efficient Separation of Eleven Mycosporine-like Amino Acids by UHPLC-DAD and Their Quantification in Diverse Red Algae
Abstract
:1. Introduction
2. Results
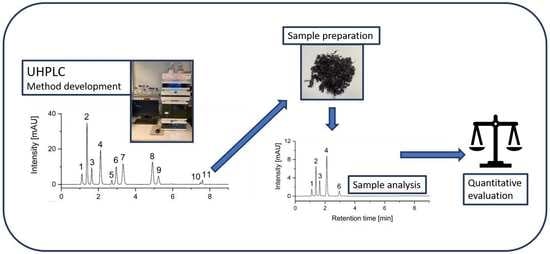
2.1. Sample Preparation
2.2. UHPLC Method Development
2.3. Method Validation
2.4. Analysis of Different Algal Species
3. Materials and Methods
3.1. Biomaterial and Preparation of Crude Extracts
3.2. Chemicals and Reagents
3.3. Analytical Conditions
3.3.1. UHPLC-DAD
3.3.2. UHPLC-MS
3.4. Calibration and Method Validation
3.4.1. Linearity, Limit of Detection (LOD) and Limit of Quantification (LOQ)
3.4.2. Precision and Accuracy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Orfanoudaki, M.; Hartmann, A.; Karsten, U.; Ganzera, M. Chemical profiling of mycosporine-like amino acids in twenty-three red algal species. J. Phycol. 2019, 55, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Becker, K.; Karsten, U.; Remias, D.; Ganzera, M. Analysis of mycosporine-like amino acids in selected algae and cyanobacteria by hydrophilic interaction liquid chromatography and a novel MAA from the red alga Catenella repens. Mar. Drugs 2015, 13, 6291–6305. [Google Scholar] [CrossRef] [PubMed]
- Cardozo, K.H.M.; Marques, L.G.; Carvalho, V.M.; Carignan, M.O.; Pinto, E.; Marinho-Soriano, E.; Colepicolo, P. Analyses of photoprotective compounds in red algae from the Brazilian coast. Rev. Bras. Farmacogn. 2011, 21, 202–208. [Google Scholar] [CrossRef] [Green Version]
- Nishida, Y.; Kumagai, Y.; Michiba, S.; Yasui, H.; Kishimura, H. Efficient extraction and antioxidant capacity of mycosporine-like amino acids from red alga dulse Palmaria palmata in Japan. Mar. Drugs 2020, 18, 502. [Google Scholar] [CrossRef]
- Ngoennet, S.; Nishikawa, Y.; Hibino, T.; Waditee-Sirisattha, R.; Kageyama, H. A method for the isolation and characterization of mycosporine-like amino acids from cyanobacteria. Methods Protoc. 2018, 1, 46. [Google Scholar] [CrossRef] [Green Version]
- Figueroa, F.L. Mycosporine-like amino acids from marine resource. Mar. Drugs 2021, 19, 18. [Google Scholar] [CrossRef]
- Suh, S.S.; Lee, S.G.; Youn, U.J.; Han, S.J.; Kim, I.C.; Kim, S. Comprehensive expression profiling and functional network analysis of Porphyra-334, one mycosporine-like amino acid (MAA), in human keratinocyte exposed with UV-radiation. Mar. Drugs 2017, 15, 196. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, A.; Holzinger, A.; Ganzera, M.; Karsten, U. Prasiolin, a new UV-sunscreen compound in the terrestrial green macroalga Prasiola calophylla (Carmichael ex Greville) Kutzing (Trebouxiophyceae, Chlorophyta). Planta 2016, 243, 161–169. [Google Scholar] [CrossRef] [Green Version]
- Conde, F.R.; Carignan, M.O.; Sandra Churio, M.; Carreto, J.I. In vitro cis–trans photoisomerization of palythene and usujirene. Implications on the in vivo transformation of mycosporine-like amino acids. Photochem. Photobiol. 2003, 77, 146–150. [Google Scholar] [CrossRef]
- Vega, J.; Schneider, G.; Moreira, B.R.; Herrera, C.; Bonomi-Barufi, J.; Figueroa, F.L. Mycosporine-like amino acids from red macroalgae: UV-photoprotectors with potential cosmeceutical applications. Appl. Sci. 2021, 11, 5112. [Google Scholar] [CrossRef]
- Sinha, R.P.; Klisch, M.; Gröniger, A.; Häder, D.-P. Mycosporine-like amino acids in the marine red alga Gracilaria cornea—Effects of UV and heat. Environ. Exp. Bot. 2000, 43, 33–43. [Google Scholar] [CrossRef]
- Wada, N.; Sakamoto, T.; Matsugo, S. Mycosporine-like amino acids and their derivatives as natural antioxidants. Antioxidants 2015, 4, 603–646. [Google Scholar] [CrossRef] [PubMed]
- Ying, R.; Zhang, Z.; Zhu, H.; Li, B.; Hou, H. The protective effect of mycosporine-like amino acids (MAAs) from Porphyra yezoensis in a mouse model of UV irradiation-induced photoaging. Mar. Drugs 2019, 17, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosic, N.N. Mycosporine-like amino acids: Making the foundation for organic personalised sunscreens. Mar. Drugs 2019, 17, 638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, D.; Schürch, C.; Zülli, F. Mycosporine-like amino acids from red algae protect against premature skin-aging. Euro Cosmet 2006, 9, 1–4. [Google Scholar]
- Corinaldesi, C.; Barone, G.; Marcellini, F.; Dell’Anno, A.; Danovaro, R. Marine microbial-derived molecules and their potential use in cosmeceutical and cosmetic products. Mar. Drugs 2017, 15, 118. [Google Scholar] [CrossRef]
- Becker, K.; Hartmann, A.; Ganzera, M.; Fuchs, D.; Gostner, J.M. Immunomodulatory effects of the mycosporine-like amino acids shinorine and porphyra-334. Mar. Drugs 2016, 14, 119. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, A.; Gostner, J.; Fuchs, J.E.; Chaita, E.; Aligiannis, N.; Skaltsounis, L.; Ganzera, M. Inhibition of collagenase by mycosporine-like amino acids from marine sources. Planta Med. 2015, 81, 813–820. [Google Scholar] [CrossRef] [Green Version]
- Ryu, J.; Park, S.J.; Kim, I.H.; Choi, Y.H.; Nam, T.J. Protective effect of porphyra-334 on UVA-induced photoaging in human skin fibroblasts. Int. J. Mol. Med. 2014, 34, 796–803. [Google Scholar] [CrossRef] [Green Version]
- Rosic, N.N. Recent advances in the discovery of novel marine natural products and mycosporine-like amino acid UV-absorbing compounds. Appl. Microbiol. Biotechnol. 2021, 105, 7053–7067. [Google Scholar] [CrossRef]
- Suh, S.S.; Hwang, J.; Park, M.; Seo, H.H.; Kim, H.S.; Lee, J.H.; Moh, S.H.; Lee, T.K. Anti-inflammation activities of mycosporine-like amino acids (MAAs) in response to UV radiation suggest potential anti-skin aging activity. Mar. Drugs 2014, 12, 5174–5187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raj, S.; Kuniyil, A.M.; Sreenikethanam, A.; Gugulothu, P.; Jeyakumar, R.B.; Bajhaiya, A.K. Microalgae as a source of mycosporine-like amino acids (MAAs); advances and future prospects. Int. J. Environ. Res. Public Health 2021, 18, 12402. [Google Scholar] [CrossRef] [PubMed]
- Parailloux, M.; Godin, S.; Fernandes, S.C.M.; Lobinski, R. Untargeted analysis for mycosporines and mycosporine-like amino acids by hydrophilic interaction liquid chromatography (HILIC)-electrospray orbitrap MS(2)/MS(3). Antioxidants 2020, 9, 1185. [Google Scholar] [CrossRef] [PubMed]
- Orfanoudaki, M.; Hartmann, A.; Mayr, J.; Figueroa, F.L.; Vega, J.; West, J.; Bermejo, R.; Maggs, C.; Ganzera, M. Analysis of the mycosporine-like amino acid (MAA) pattern of the salt marsh red alga Bostrychia scorpioides. Mar. Drugs 2021, 19, 321. [Google Scholar] [CrossRef] [PubMed]
- Gambichler, V.; Zuccarello, G.C.; Karsten, U. Seasonal changes in stress metabolites of native and introduced red algae in New Zealand. J. Appl. Phycol. 2021, 33, 1157–1170. [Google Scholar] [CrossRef]
- Orfanoudaki, M.; Hartmann, A.; Kamiya, M.; West, J.; Ganzera, M. Chemotaxonomic study of Bostrychia spp. (Ceramiales, Rhodophyta) based on their mycosporine-like amino acid content. Molecules 2020, 25, 3273. [Google Scholar] [CrossRef]
- Chaves-Pena, P.; de la Coba, F.; Figueroa, F.L.; Korbee, N. Quantitative and qualitative HPLC analysis of mycosporine-like amino acids extracted in distilled water for cosmetical uses in four rhodophyta. Mar. Drugs 2019, 18, 27. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, A.; Murauer, A.; Ganzera, M. Quantitative analysis of mycosporine-like amino acids in marine algae by capillary electrophoresis with diode-array detection. J. Pharm. Biomed. Anal. 2017, 138, 153–157. [Google Scholar] [CrossRef]
- Geraldes, V.; Jacinavicius, F.R.; Genuario, D.B.; Pinto, E. Identification and distribution of mycosporine-like amino acids in Brazilian cyanobacteria using ultrahigh-performance liquid chromatography with diode array detection coupled to quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass. Spectrom. 2020, 34 (Suppl. S3), e8634. [Google Scholar] [CrossRef]
- Karsten, U.; Escoubeyrou, K.; Charles, F. The effect of re-dissolution solvents and HPLC columns on the analysis of mycosporine-like amino acids in the eulittoral macroalgae Prasiola crispa and Porphyra umbilicalis. Helgol. Mar. Res. 2009, 63, 231–238. [Google Scholar] [CrossRef] [Green Version]
- ICH. Available online: https://www.ich.org/ (accessed on 17 January 2022).


| Calibration Data | |||||
|---|---|---|---|---|---|
| Compound | Regression Equation | R2 | Linear Range * | LOD * | LOQ * |
| 1 | y = 26.408x + 1.6024 | 0.9999 | 0.06–124 | 0.01 | 0.04 |
| 2 | y = 14.121x + 2.0619 | 0.9998 | 0.05–205 | 0.01 | 0.03 |
| 3 | y = 18.850x + 2.0478 | 0.9998 | 0.05–105 | 0.01 | 0.04 |
| 4 | y = 26.120x +2.3516 | 0.9999 | 0.06–113 | 0.01 | 0.04 |
| 6 | y = 14.598x +2.5545 | 0.9999 | 0.08–163 | 0.01 | 0.03 |
| 7 | y = 18.789x +2.1740 | 0.9999 | 0.06–128 | 0.01 | 0.03 |
| 9 | y = 11.766x +1.9540 | 0.9998 | 0.08–172 | 0.01 | 0.04 |
| Compound | Relative Standard Deviation | |||
|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Inter-Day | |
| 1 | 2.68 | 2.16 | 2.62 | 2.05 |
| 2 | 2.67 | 2.50 | 2.77 | 0.71 |
| 3 | 1.11 | 2.33 | 2.54 | 2.42 |
| 4 | 1.51 | 1.71 | 1.61 | 1.38 |
| 6 | 2.12 | 1.99 | 1.82 | 2.53 |
| Compound | Recovery Rate (%) | ||
|---|---|---|---|
| Low | Medium | Low | |
| 1 | 101.8 (1.1) | 103.4 (1.3) | 100.1 (1.2) |
| 2 | 97.5 (0.7) | 102.3 (1.1) | 102.2 (2.3) |
| 3 | 100.8 (1.1) | 102.9 (0.8) | 102.8 (2.3) |
| 4 | 98.2 (1.8) | 101.6 (1.0) | 100.3 (1.5) |
| 6 | 103.2 (1.8) | 97.3 (0.6) | 103.3 (2.2) |
| 7 | 100.4 (2.6) | 102.4 (1.7) | 98.8 (1.6) |
| 9 | 101.1 (1.8) | 97.9 (1.6) | 97.4 (1.9) |
| Compound | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alga | ||||||||||||
| Caloglossa ogasawaraensis | 0.13 (1.57) | 3.41 (0.60) | - | - | - | - | - | - | - | |||
| Ceramium sp. (a) | 0.10 (1.02) | 0.04 (1.81) | Det | 1.31 (0.71) | Det | 0.04 (2.02) | Det | Det | Det | - | - | |
| Ceramium sp. (b) | 1.13 (2.59) | 0.54 (1.26) | - | 0.06 (1.95) | - | - | - | - | - | - | - | |
| Chondrus crispus | 0.28 (2.11) | 1.36 (1.51) | 0.38 (1.48) | - | - | - | - | - | - | - | - | |
| Euptilorta formosissima | Det | 0.02 (2.47) | Det | Det | - | Det | Det | - | - | - | - | |
| Gracilaria chilensis | 0.40 (0.47) | 1.30 (1.03) | 0.35 (0.57) | 2.04 (1.17) | Det | 0.47 (0.96) | - | - | - | Det | Det | |
| Gracilaria gracilis | 3.54 (1.29) | - | - | 0.03 (2.48) | - | - | - | - | - | - | - | |
| Grateolupia turuturu | 2.42 (1.81) | 1.06 (2.56) | 0.04 (1.00) | - | - | - | - | - | - | - | - | |
| Jania rubens (a) | 0.16 (2.54) | 0.08 (1.28) | - | - | - | - | - | - | - | - | - | |
| Jania rubens (b) | 0.05 (1.92) | 0.16 (1.14) | - | - | - | - | - | - | - | - | - | |
| Mastocarpus stellatus | 2.83 (0.69) | 0.29 (1.66) | 0.06 (1.51) | 0.05 (2.40) | - | - | - | - | - | - | - | |
| Osmundea sp. | 0.09 (1.07) | 0.51 (1.34) | 0.27 (1.70) | 0.69 (2.03) | - | 0.06 (1.28) | - | - | - | - | - | |
| Porphyra columbina | 1.07 (0.46) | 0.11 (0.99) | Det | 2.51 (0.65) | 0.07 (1.81) | - | 0.17 (0.63) | - | - | - | - | |
| Porphyra sp. (Nori a) | 0.17 (0.82) | 0.36 (1.30) | 0.45 (1.15) | 6.58 (1.17) | - | - | - | - | - | - | - | |
| Porphyra sp. (Nori b) | 0.21 (0.52) | 0.13 (1.34) | 0.27 (0.64) | 4.36 (1.94) | - | - | - | - | - | - | - | |
| Porphyra sp. (Nori c) | 4.14 (0.52) | 0.17 (2.46) | - | 5.62 (1.92) | - | - | - | - | - | - | - | |
| Pterocladia sp. | 0.20 (0.63) | Det | - | Det | - | - | - | - | - | - | - | |
| Pyropia plicata (a) | 2.38 (0.40) | 1.52 (0.15) | Det | 3.82 (0.41) | - | 0.05 (1.46) | 0.38 (1.12) | - | 0.10 (1.92) | - | - | |
| Pyropia plicata (b) | 1.60 (0.83) | 0.77 (1.41) | Det | 2.40 (0.51) | Det | 0.05 (1.51) | 0.11 (0.89) | - | - | Det | Det | |
| Pyropia plicata (c) | 1.24 (0.50) | 0.33 (1.79) | Det | 2.13 (0.56) | - | 0.03 (1.99) | 0.13 (0.40) | - | - | Det | Det | |
| Pyropia umbilicalis | 0.77 (0.56) | 0.76 (1.17) | 0.13 (1.69) | 10.21 (0.24) | - | 0.62 (0.73) | 0.11 (1.51) | - | - | - | - | |
| Schizymenia apoda | 0.04 (0.88) | 0.02 (2.81) | - | Det | - | - | - | - | - | - | - | |
| Spongoclonium pastorale | 0.10 (0.37) | Det | - | 3.51 (0.36) | - | - | - | - | - | - | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zwerger, M.; Ganzera, M. Fast and Efficient Separation of Eleven Mycosporine-like Amino Acids by UHPLC-DAD and Their Quantification in Diverse Red Algae. Mar. Drugs 2022, 20, 395. https://doi.org/10.3390/md20060395
Zwerger M, Ganzera M. Fast and Efficient Separation of Eleven Mycosporine-like Amino Acids by UHPLC-DAD and Their Quantification in Diverse Red Algae. Marine Drugs. 2022; 20(6):395. https://doi.org/10.3390/md20060395
Chicago/Turabian StyleZwerger, Michael, and Markus Ganzera. 2022. "Fast and Efficient Separation of Eleven Mycosporine-like Amino Acids by UHPLC-DAD and Their Quantification in Diverse Red Algae" Marine Drugs 20, no. 6: 395. https://doi.org/10.3390/md20060395