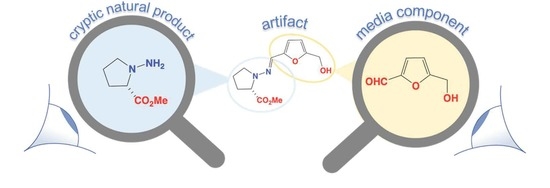
N-Amino-l-Proline Methyl Ester from an Australian Fish Gut-Derived Fungus: Challenging the Distinction between Natural Product and Artifact
Abstract
:1. Introduction
2. Results
2.1. Uninoculated Rice Media Is the Source of Furan Aldehydes
2.2. Detection of N-Amino-l-Proline Methyl Ester (5) and Its Free Carboxylic Acid 6 Using 2,4-Dinitrobenzaldehyde (2,4-DNB) as an In Situ Derivatizing Agent
2.3. N-Amino-l-Proline Methyl Ester (5) (but Not the Free Carboxylic Acid 6) in CMB-F563 Rice Grain Media Cultivations
2.4. Furans 7–8 in Uninoculated M1 Broth Media
2.5. N-Amino-l-Proline Methyl Ester (5) in CMB-F563 M1 Broth Media Cultivations
2.6. To Secrete or Not Secrete N-Amino-l-Proline Methyl Ester (5)
2.7. N-Amino-l-Proline Methyl Ester (5) in Other CMB-F563 Broth Media Cultivations
2.8. N-Amino-l-Proline Methyl Ester (5) in Broth Media Cultivations of Other Fungi
3. Discussion
3.1. Origins of Prolinimines 1–4, N-Amino-l-Proline Methyl Ester (5) and Furans 7–8
- (i)
- As the furans 7–8 are generated in the rice grain media during autoclaving, prior to fungal inoculation, they are best designated as media constituents.
- (ii)
- CMB-F563 cultivations that produce the cryptic natural product N-amino-l-proline methyl ester (5), retain it exclusively within fungal mycelia.
- (iii)
- The rupture of CMB-F563 mycelia during solvent extraction allow 5 to come in contact with media constituent furans 7–8, facilitating rapid in situ formation of the Schiff base prolinimines A–B (1–2). As 1–2 are produced as a direct result of the extraction process they are more aptly characterized as artifacts.
- (iv)
- During fractionation/handling prolinimine A (1) undergoes acid-mediated conversion to prolinimines C–D (3–4), consistent with 3–4 being artifacts.
- (v)
- Autoclaving media with low carbohydrate content (i.e., M1) fails to produce significant levels of the furans 7–8.
- (vi)
- Those CMB-F563 cultivations that produce the natural product 5, but lack the media components 7–8 (i.e., M1), are incapable of forming the artifacts 1–4.
- (vii)
- As 5 has cryptic physical and spectroscopic properties that make it hard to detect in cultures/extracts, it easily overlooked (unless you know what you are looking for, and apply appropriate measures—i.e., 2,4-DNB derivatization).
- (viii)
- Notwithstanding, as 5 can be readily detected in culture/extracts by in situ chemical derivatization with 2,4-DNB to form the Schiff base 9, it can be designated as a natural product.
3.2. Knowledge of N-Amino-Prolines in Nature
- Is flaxseed the source of linatine (12), or is 12 a fungal natural product? If the latter can flaxseed meal be treated to render it suitable as a chick feed?
- What is the ecological (survival) advantage accrued to CMB-F563 by its ability to produce 5, and is there any correlation between this and the isolation of CMB-F563 from a fish gastrointestinal tract?
- Given the cryptic nature of 5, how common is it (or other N-amino amino acids) as a fungal natural product?
4. Materials and Methods
4.1. General Experimental Details
4.2. Collection and Taxonomy of Evlachovaea sp. CMB-F563
4.3. Synthesis of Authentic Standards
4.3.1. Synthesis of N-Amino-l-Proline Methyl Ester (5)
4.3.2. Synthesis of 2,4-Dinitrobenzaldehyde Schiff Base 9
4.3.3. Synthesis of 2,4-Dinitrobenzaldehyde Schiff Base 10
4.4. The Presence of Furans 7–8, and Absence of 5, in Uninoculated Rice Grain Media
4.5. Detection of N-Aminoprolines 5–6 Using 2,4-Dinitrobenzaldehyde
4.6. Detection of N-Amino-l-Proline Methyl Ester (5) in CMB-F563 Rice Grain Media Cultivations
4.7. Detection of N-Amino-l-Proline Methyl Ester (5) in Uninoculated M1 Broth Media
4.8. Detection of N-Amino-l-Proline Methyl Ester (5) in CMB-F563 M1 Broth Media Cultivations
4.9. Mycelial Secretion Status of N-Amino-l-Proline Methyl Ester (5)
4.9.1. Using 2,4-Dinitrobenzaldehyde (2,4-DNB) as the Derivatizing Agent
4.9.2. Using 5-Hydroxymethyfurfural (7) as the Derivatizing Agent
4.10. Detection of N-Amino-l-Proline Methyl Ester (5) in Other CMB-F563 Broth Media Cultivations
4.11. Detection of N-Amino-l-Proline Methyl Ester (5) in Different Fungal Genera
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capon, R.J. Extracting value: Mechanistic insights into the formation of natural product artifacts—Case studies in marine natural products. Nat. Prod. Rep. 2020, 37, 55–79. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, O.G.; Khalil, Z.G.; Capon, R.J. Prolinimines: N-Amino-l-Pro-methyl Ester (Hydrazine) Schiff Bases from a Fish Gastrointestinal Tract-Derived Fungus, Trichoderma sp. CMB-F563. Org. Lett. 2018, 20, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huang, H.; Liu, W.; Peng, N.; Huang, X. Kinetics of the 5-Hydroxymethylfurfural Formation Reaction in Chinese Rice Wine. J. Agric. Food Chem. 2010, 58, 3507–3511. [Google Scholar] [CrossRef] [PubMed]
- Shapla, U.M.; Solayman, M.; Alam, N.; Khalil, M.I.; Gan, S.H. 5-Hydroxymethylfurfural (HMF) levels in honey and other food products: Effects on bees and human health. Chem. Cent. J. 2018, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Abraham, K.; Guertler, R.; Berg, K.; Heinemeyer, G.; Lampen, A.; Appel, K.E. Toxicology and risk assessment of 5-Hydroxymethylfurfural in food. Mol. Nutr. Food Res. 2011, 55, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Capuano, E.; Fogliano, V. Acrylamide and 5-hydroxymethylfurfural (HMF): A review on metabolism, toxicity, occurrence in food and mitigation strategies. LWT—Food Sci. Technol. 2011, 44, 793–810. [Google Scholar] [CrossRef]
- Mohamed, O.G.; Khalil, Z.G.; Salim, A.A.; Cui, H.; Blumenthal, A.; Capon, R.J. Lincolnenins A–D: Isomeric bactericidal bianthracenes from Streptomyces lincolnensis. J. Org. Chem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Khalil, Z.; Li, L.; Salim, A.A.; Quezada, M.; Kalansuriya, P.; Capon, R. Roseopurpurins: Chemical Diversity Enhanced by Convergent Biosynthesis and Forward and Reverse Michael Additions. J. Org. Lett. 2016, 18, 4340–4343. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Li, L.; Espósito, B.P.; Salim, A.A.; Khalil, Z.G.; Quezada, M.; Bernhardt, P.V.; Capon, R.J. New PKS-NRPS tetramic acids and pyridinone from an Australian marine-derived fungus, Chaunopycnis sp. Org. Biomol. Chem. 2015, 13, 7795–7802. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Salim, A.A.; Khalil, Z.; Quezada, M.; Bernhardt, P.V.; Capon, R.J. Viridicatumtoxins: Expanding on a Rare Tetracycline Antibiotic Scaffold. J. Org. Chem. 2015, 80, 12501–12508. [Google Scholar] [CrossRef] [PubMed]
- Klosterman, H.J.; Lamoureux, G.L.; Parsons, J.L. Isolation, characterization, and synthesis of linatine. A vitamin B6 antagonist from flaxseed (Linum usitatissimum). Biochemistry 1967, 6, 170–177. [Google Scholar] [CrossRef]
- Bekhit, A.E.D.A.; Shavandi, A.; Jodjaja, T.; Birch, J.; Teh, S.; Ahmed, I.A.M.; Al-Juhaimi, F.Y.; Saeedi, P.; Bekhit, A.A. Flaxseed: Composition, detoxification, utilization, and opportunities. Biocatal. Agric. Biotechnol. 2018, 13, 129–152. [Google Scholar] [CrossRef]
- Mayengbam, S.; Yang, H.; Barthet, V.; Aliani, M.; House, J.D. Identification, Characterization, and Quantification of an Anti-pyridoxine Factor from Flaxseed Using Ultrahigh-Performance Liquid Chromatography–Mass Spectrometry. J. Agric. Food Chem. 2014, 62, 419–426. [Google Scholar] [CrossRef] [PubMed]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, O.G.; Khalil, Z.G.; Capon, R.J. N-Amino-l-Proline Methyl Ester from an Australian Fish Gut-Derived Fungus: Challenging the Distinction between Natural Product and Artifact. Mar. Drugs 2021, 19, 151. https://doi.org/10.3390/md19030151
Mohamed OG, Khalil ZG, Capon RJ. N-Amino-l-Proline Methyl Ester from an Australian Fish Gut-Derived Fungus: Challenging the Distinction between Natural Product and Artifact. Marine Drugs. 2021; 19(3):151. https://doi.org/10.3390/md19030151
Chicago/Turabian StyleMohamed, Osama G., Zeinab G. Khalil, and Robert J. Capon. 2021. "N-Amino-l-Proline Methyl Ester from an Australian Fish Gut-Derived Fungus: Challenging the Distinction between Natural Product and Artifact" Marine Drugs 19, no. 3: 151. https://doi.org/10.3390/md19030151