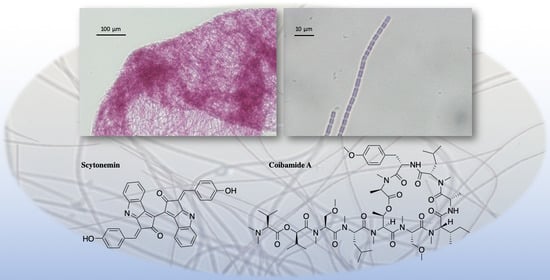
The Chemistry, Biochemistry and Pharmacology of Marine Natural Products from Leptolyngbya, a Chemically Endowed Genus of Cyanobacteria
Abstract
:1. Introduction
2. Chemical Diversity of the Secondary Metabolites Isolated from Leptolyngbya
2.1. Polypeptides
2.2. Simple Esters
2.3. Macrolides
2.4. Pyrones
2.5. Polyaromatics
2.6. Oxazolines
2.7. Other
2.7.1. Toxins
2.7.2. Non-Toxic Metabolites
2.7.3. Phenolic Compounds
2.7.4. Odorous Metabolites
2.7.5. Pigments
3. Laboratory Cultivation
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gademann, K.; Portmann, C. Secondary metabolites from cyanobacteria: Complex structures and powerful bioactivities. Curr. Org. Chem. 2008, 12, 326–341. [Google Scholar] [CrossRef]
- Summons, R.E.; Jahnke, L.L.; Hope, J.M.; Logan, G.A. 2-Methylhopanoids as biomarkers forcyanobacterial oxygenic photosynthesis. Nature 1999, 400, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Blankenship, R.E.; Hartman, H. The origin and evolution of oxygenic photosynthesis. Trends Biochem. Sci. 1998, 23, 94–97. [Google Scholar] [CrossRef]
- Berman-Frank, I.; Lundgren, P.; Falkowski, P. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 2003, 154, 157–164. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, W.; Jeon, S.M.; Kim, T.; Park, A.; Kim, J.; Heo, S.J.; Oh, C.; Shim, W.B.; Kang, D.H. Isolation and characterization of Leptolyngbya sp. KIOST-1, a basophilic and euryhaline filamentous cyanobacterium from an open paddle-wheel raceway Arthrospira culture pond in Korea. J. Appl. Microbiol. 2015, 119, 1597–1612. [Google Scholar] [CrossRef] [Green Version]
- Mai, T.; Johansen, J.R.; Pietrasiak, N.; Bohunická, M.; Martin, M.P. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 2018, 365, 1–59. [Google Scholar] [CrossRef] [Green Version]
- Bruno, L.; Billi, D.; Bellezza, S.; Albertano, P. Cytomorphological and genetic characterization of troglobitic Leptolyngbya strains isolated from roman hypogea. Appl. Environ. Microbiol. 2009, 75, 608–617. [Google Scholar] [CrossRef] [Green Version]
- Anagnostidis, K.; Komárek, J. Modern approach to the classification system of cyanophytes. 3—Oscillatoriales. Algol. Stud. /Arch. Hydrobiol. Suppl. Vol. 1988, 50–53, 327–472. [Google Scholar]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Cirés, S.; Casero, M.C.; Quesada, A. Toxicity at the edge of life: A review on cyanobacterial toxins from extreme environments. Mar. Drugs 2017, 15, 233. [Google Scholar] [CrossRef]
- Ahmed, M.; Stal, L.J.; Hasnain, S. The morphology and bioactivity of the rice field cyanobacterium Leptolyngbya. Rev. Biol. Trop. 2014, 62, 1251–1260. [Google Scholar] [PubMed]
- Veerabadhran, M.; Chakraborty, S.; Mitra, S.; Karmakar, S.; Mukherjee, J. Effects of flask configuration on biofilm growth and metabolites of intertidal cyanobacteria isolated from a mangrove forest. J. Appl. Microbiol. 2018, 125, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, Z.A.; Al-Shehri, A.M. Biodiversity and toxin production of cyanobacteria in mangrove swamps in the Red Sea off the southern coast of Saudi Arabia. Bot. Mar. 2015, 58, 23–34. [Google Scholar] [CrossRef]
- Yuu, H.; Fujisawa, T.; Ohtsubo, Y.; Katayama, M.; Misawa, N.; Wakazuki, S.; Shimura, Y.; Nakamura, Y.; Kawachi, M.; Yoshikawa, H.; et al. Complete genome sequence of cyanobacterium Leptolyngbya sp. NIES-3755. Am. Soc. Microbiol. 2016, 4, e0009-16. [Google Scholar]
- Brito, Â.; Ramos, V.; Seabra, R.; Santos, A.; Santos, C.L.; Lopo, M.; Ferreira, S.; Martins, A.; Mota, R.; Frazão, B.; et al. Culture-dependent characterization of cyanobacterial diversity in the intertidal zones of the Portuguese coast: A polyphasic study. Syst. Appl. Microbiol. 2012, 35, 110–119. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.; Garcia, M.; Costa-Rpdrigues, J.; Costa, M.S.; Ribeiro, M.J.; Fernandes, M.H.; Barros, P.; Barreiro, A.; Vasconcelos, V.; Martins, R. Exploring bioactive properties of marine cyanobacteria isolated from the Portuguese Coast: High potential as a source of anticancer compounds. Mar. Drugs 2014, 12, 98–114. [Google Scholar] [CrossRef] [Green Version]
- Semary, N.A. El Microscopic, molecular, and biochemical investigations to characterize a benthic cyanoprokaryote Leptolyngbya strain from Egypt. Microsc. Res. Tech. 2013, 76, 249–257. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. Available online: https://www.algaebase.org (accessed on 3 October 2020).
- Komárek, J.; Hauer, T. The On-line Database of Cyanobacterial Genera. Available online: http://www.cyanodb.cz (accessed on 3 October 2020).
- Singh, D.P.; Khattar, J.I.S.; Gupta, M.; Kaur, G. Evaluation of toxicological impact of cartap hydrochloride on some physiological activities of a non-heterocystous cyanobacterium Leptolyngbya foveolarum. Pestic. Biochem. Physiol. 2014, 110, 63–70. [Google Scholar] [CrossRef]
- Pramanik, A.; Sundararaman, M.; Das, S.; Ghosh, U.; Mukherjee, J. Isolation and characterization of cyanobacteria possessing antimicrobial activity from the sundarbans, the world’s largest tidal mangrove forest. J. Phycol. 2011, 47, 731–743. [Google Scholar] [CrossRef]
- Xue, Y.; Zhao, P.; Quan, C.; Zhao, Z.; Gao, W.; Li, J.; Zu, X.; Fu, D.; Feng, S.; Bai, X.; et al. Cyanobacteria-derived peptide antibiotics discovered since 2000. Peptides 2018, 107, 17–24. [Google Scholar] [CrossRef]
- Haque, F.; Banayan, S.; Yee, J.; Chiang, Y.W. Extraction and applications of cyanotoxins and other cyanobacterial secondary metabolites. Chemosphere 2017, 183, 164–175. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Menakha, M. Pharmaceutical applications of cyanobacteria-A review. J. Acute Med. 2015, 5, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Engene, N.; Gunasekera, S.P.; Gerwick, W.H.; Paul, V.J. Phylogenetic inferences reveal a large extent of novel biodiversity in chemically rich tropical marine cyanobacteria. Appl. Environ. Microbiol. 2013, 79, 1882–1888. [Google Scholar] [CrossRef] [Green Version]
- Bertin, M.J.; Vulpanovici, A.; Monroe, E.A.; Korobeynikov, A.; Sherman, D.H.; Gerwick, L.; Gerwick, W.H. The Phormidolide biosynthetic gene cluster: A trans-AT PKS pathway encoding a toxic macrocyclic polyketide. ChemBioChem 2016, 17, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Castenholz, R.W. Culturing methods for cyanobacteria. Methods Enzymol. 1988, 167, 68–93. [Google Scholar]
- Moss, N.A.; Leao, T.; Glukhov, E.; Gerwick, L.; Gerwick, W.H. Collection, Culturing, and Genome Analyses of Tropical Marine Filamentous Benthic Cyanobacteria, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 604, ISBN 9780128139592. [Google Scholar]
- Leão, P.N.; Ramos, V.; Goncalves, P.B.; Viana, F.; Lage, O.M.; Gerwick, W.H.; Vasconcelos, V.M. Chemoecological screening reveals high bioactivity in diverse culturable portuguese marine cyanobacteria. Mar. Drugs 2013, 11, 1316–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, G.; Pan, Z.; Wu, C.; Wang, W.; Fang, L.; Su, W. Efficient synthesis and stereochemical revision of Coibamide, A. J. Am. Chem. Soc. 2015, 137, 13488–13491. [Google Scholar] [CrossRef]
- Yao, G.; Wang, W.; Ao, L.; Cheng, Z.; Wu, C.; Pan, Z.; Liu, K.; Li, H.; Su, W.; Fang, L. Improved total synthesis and biological evaluation of coibamide A analogues. J. Med. Chem. 2018, 61, 8908–8916. [Google Scholar] [CrossRef]
- Medina, R.A.; Goeger, D.E.; Hills, P.; Mooberry, S.L.; Huang, N.; Romero, L.I.; Ortega-Barría, E.; Gerwick, W.H.; McPhail, K.L. Coibamide A, a potent antiproliferative cyclic depsipeptide from the panamanian marine cyanobacterium Leptolyngbya sp. J. Am. Chem. Soc. 2008, 130, 6324–6325. [Google Scholar] [CrossRef] [Green Version]
- Sable, G.A.; Park, J.; Kim, H.; Lim, S.J.; Jang, S.; Lim, D. Solid-Phase Total synthesis of the proposed structure of coibamide A and its derivative: Highly methylated cyclic depsipeptides. Eur. J. Org. Chem. 2015, 2015, 7043–7052. [Google Scholar] [CrossRef]
- He, W.; Qiu, H.B.; Chen, Y.J.; Xi, J.; Yao, Z.J. Total synthesis of proposed structure of coibamide A, a highly N- and O-methylated cytotoxic marine cyclodepsipeptide. Tetrahedron Lett. 2014, 55, 6109–6112. [Google Scholar] [CrossRef]
- Nabika, R.; Suyama, T.L.; Hau, A.M.; Misu, R.; Ohno, H.; Ishmael, J.E.; McPhail, K.L.; Oishi, S.; Fujii, N. Synthesis and biological evaluation of the [d-MeAla11]-epimer of coibamide A. Bioorg. Med. Chem. Lett. 2015, 25, 302–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thornburg, C.C.; Thimmaiah, M.; Shaala, L.A.; Hau, A.M.; Malmo, J.M.; Ishmael, J.E.; Youssef, D.T.A.; McPhail, K.L. Cyclic depsipeptides, grassypeptolides D and E and Ibu- epidemethoxylyngbyastatin 3, from a Red Sea Leptolyngbya cyanobacterium. J. Nat. Prod. 2011, 74, 1677–1685. [Google Scholar] [CrossRef] [Green Version]
- Harrigan, G.G.; Yoshida, W.Y.; Moore, R.E.; Nagle, D.G.; Park, P.U.; Biggs, J.; Paul, V.J.; Mooberry, S.L.; Corbett, T.H.; Valeriote, F.A. Isolation, structure determination, and biological activity of dolastatin 12 and lyngbyastatin 1 from Lyngbya majuscula/Schizothrix calcicola cyanobacterial assemblages. J. Nat. Prod. 1998, 61, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Pettit, G.R.; Kamano, Y.; Kizu, H.; Dufresne, C.; Herald, C.L.; Bontems, R.J.; Schmidt, J.M.; Boettner, F.E.; Nieman, R.A. Isolation and structure of the cell growth inhibitory depsipeptides dolastatins 11 and 12. Heterocycles 1989, 28, 3–8. [Google Scholar] [CrossRef]
- Bates, R.B.; Brusoe, K.G.; Burns, J.J.; Caldera, S.; Cui, W.; Gangwar, S.; Gramme, M.R.; McClure, K.J.; Rouen, G.P.; Schadow, H.; et al. Dolastatins. 26. synthesis and stereochemistry of dolastatin 11. J. Am. Chem. Soc. 1997, 119, 2111–2113. [Google Scholar] [CrossRef]
- Al-Awadhi, F.H.; Paul, V.J.; Luesch, H. Structural diversity and anticancer activity of marine-derived elastase inhibitors: Key features and mechanisms mediating the antimetastatic effects in invasive breast cancer. ChemBioChem 2018, 19, 815–825. [Google Scholar] [CrossRef]
- Maneechote, N.; Yingyongnarongkul, B.E.; Suksamran, A.; Lumyong, S. Inhibition of Vibrio spp. by 2-Hydroxyethyl-11-hydroxyhexadec-9-enoate of marine cyanobacterium Leptolyngbya sp. LT19. Aquac. Res. 2017, 48, 2088–2095. [Google Scholar] [CrossRef]
- Choi, H.; Mascuch, S.J.; Villa, F.A.; Byrum, T.; Teasdale, M.E.; Smith, J.E.; Preskitt, L.B.; Rowley, D.C.; Gerwick, L.; Gerwick, W.H. Honaucins A-C, potent inhibitors of inflammation and bacterial quorum sensing: Synthetic derivatives and structure-activity relationships. Chem. Biol. 2012, 19, 589–598. [Google Scholar] [CrossRef] [Green Version]
- Sapkota, M.; Li, L.; Choi, H.; Gerwick, W.H.; Soh, Y. Bromo-honaucin A inhibits osteoclastogenic differentiation in RAW 264.7 cells via Akt and ERK signaling pathways. Eur. J. Pharmacol. 2015, 769, 100–109. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Morita, M.; Ohno, O.; Kimura, T.; Teruya, T.; Watanabe, T.; Suenaga, K.; Shibasaki, M. Leptolyngbyolides, cytotoxic macrolides from the marine cyanobacterium Leptolyngbya sp.: Isolation, biological activity, and catalytic asymmetric total synthesis. Chem. A Eur. J. 2017, 23, 8500–8509. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.R.; Cao, Z.; Engene, N.; Soria-Mercado, I.E.; Murray, T.F.; Gerwick, W.H. Palmyrolide A, an unusually stabilized neuroactive macrolide from palmyra atoll cyanobacteria. Org. Lett. 2010, 12, 4490–4493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, R.T.; Boulanger, A.; Vulpanovici, A.; Roberts, M.A.; Gerwick, W.H. Structure and absolute stereochemistry of phormidolide, a new toxic metabolite from the marine cyanobacterium Phormidium sp. J. Org. Chem. 2002, 67, 7927–7936. [Google Scholar] [CrossRef] [PubMed]
- Bertin, M.J.; Demirkiran, O.; Navarro, G.; Moss, N.A.; Lee, J.; Goldgof, G.M.; Vigil, E.; Winzeler, E.A.; Valeriote, F.A.; Gerwick, W.H. Kalkipyrone B, a marine cyanobacterial γ-pyrone possessing cytotoxic and anti-fungal activities. Phytochemistry 2016, 122, 113–118. [Google Scholar] [CrossRef] [Green Version]
- Graber, M.A.; Gerwick, W.H. Kalkipyrone, a toxic γ-pyrone from an assemblage of the marine cyanobacteria Lyngbya majuscula and Tolypothrix sp. J. Nat. Prod. 1998, 61, 677–680. [Google Scholar] [CrossRef] [PubMed]
- Inuzuka, T.; Yamamoto, K.; Iwasaki, A.; Ohno, O.; Suenaga, K.; Kawazoe, Y.; Uemura, D. An inhibitor of the adipogenic differentiation of 3T3-L1 cells, yoshinone A, and its analogs, isolated from the marine cyanobacterium Leptolyngbya sp. Tetrahedron Lett. 2014, 55, 6711–6714. [Google Scholar] [CrossRef]
- Choi, H.; Engene, N.; Smith, J.E.; Preskitt, L.B.; Gerwick, W.H. Crossbyanols A-D, toxic brominated polyphenyl ethers from the hawai’ian bloom-forming cyanobacterium Leptolyngbya crossbyana. J. Nat. Prod. 2010, 73, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Bhandari Neupane, J.; Neupane, R.P.; Luo, Y.; Yoshida, W.Y.; Sun, R.; Williams, P.G. Characterization of leptazolines A-D, polar oxazolines from the cyanobacterium Leptolyngbya sp., reveals a glitch with the “willoughby-hoye” scripts for calculating NMR chemical shifts. Org. Lett. 2019, 21, 8449–8453. [Google Scholar] [CrossRef]
- Hau, A.M.; Greenwood, J.A.; Löhr, C.V.; Serrill, J.D.; Proteau, P.J.; Ganley, I.G.; McPhail, K.L.; Ishmael, J.E. Coibamide A induces mTOR-independent autophagy and cell death in human glioblastoma cells. PLoS ONE 2013, 8, e65250. [Google Scholar] [CrossRef] [Green Version]
- Snyder, K.M.; Sikorska, J.; Ye, T.; Fang, L.; Su, W.; Carter, R.G.; McPhail, K.L.; Cheong, P.H.Y. Towards theory driven structure elucidation of complex natural products: Mandelalides and coibamide A. Org. Biomol. Chem. 2016, 14, 5826–5831. [Google Scholar] [CrossRef]
- Pettit, G.R.; Kamano, Y.; Fujii, Y.; Herald, C.L.; Inoue, M.; Brown, P.; Gust, D.; Kitahara, K.; Schmidt, J.M.; Michael, C.; et al. Marine animal biosynthetic constituents for cancer chemotherapy. J. Nat. Prod. 1981, 44, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.; Pörzgen, P.; Pittman, E.; Yoshida, W.Y.; Westenburg, H.E.; Horgen, F.D. Isolation and structure determination of malevamide E, a dolastatin 14 analogue, from the marine cyanobacterium Symploca laete-viridis. J. Nat. Prod. 2008, 71, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Thornburg, C.C.; Mcphail, K.L. Investigation of Unique Marine Environments for Microbial Natural Products; Oregon State University: Corvallis, OR, USA, 2013. [Google Scholar]
- Williams, P.G.; Moore, R.E.; Paul, V.J. Isolation and structure determination of lyngbyastatin 3, a lyngbyastatin 1 homologue from the marine cyanobacterium Lyngbya majuscula. Determination of the configuration of the 4-Amino-2,2-dimethyl-3-oxopentanoic acid unit in majusculamide C, dolastatin 12, lyngbyastatin 1, and lyngbyastatin 3 from cyanobacteria. J. Nat. Prod. 2003, 66, 1356–1363. [Google Scholar] [PubMed]
- Gerwick, W.H.; Tan, L.T.; Sitachitta, N. Nitrogen-Containing Metabolites Marine Cyanobacteria; Elsevier: Amsterdam, The Netherlands, 2001; Volume 57. [Google Scholar]
- Kwan, J.C.; Ratnayake, R.; Abboud, K.A.; Paul, V.J.; Luesch, H. Grassypeptolides A-C, cytotoxic bis-thiazoline containing marine cyclodepsipeptides. J. Org. Chem. 2010, 75, 8012–8023. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.C.; Rocca, J.R.; Abboud, K.A.; Paul, V.J.; Luesch, H. Total structure determination of grassypeptolide, a new marine cyanobacterial cytotoxin. Org. Lett. 2008, 10, 789–792. [Google Scholar] [CrossRef]
- Gogineni, V.; Hamann, M.T. Marine natural product peptides with therapeutic potential: Chemistry, biosynthesis, and pharmacology. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 81–196. [Google Scholar] [CrossRef]
- Popplewell, W.L.; Ratnayake, R.; Wilson, J.A.; Beutler, J.A.; Colburn, N.H.; Henrich, C.J.; McMahon, J.B.; McKee, T.C. Grassypeptolides F and G, cyanobacterial peptides from Lyngbya majuscula. J. Nat. Prod. 2011, 74, 1686–1691. [Google Scholar] [CrossRef] [Green Version]
- Kwan, J.C.; Liu, Y.; Ratnayake, R.; Hatano, R.; Kuribara, A.; Morimoto, C.; Ohnuma, K.; Paul, V.J.; Ye, T.; Luesch, H. Grassypeptolides as natural inhibitors of dipeptidyl peptidase 8 and T-cell activation. ChemBioChem 2014, 15, 799–804. [Google Scholar] [CrossRef]
- Mascuch, S.J.; Boudreau, P.D.; Carland, T.M.; Pierce, N.T.; Olson, J.; Hensler, M.E.; Choi, H.; Campanale, J.; Hamdoun, A.; Nizet, V.; et al. Marine natural product honaucin A attenuates inflammation by activating the Nrf2-ARE pathway. J. Nat. Prod. 2018, 81, 506–514. [Google Scholar] [CrossRef]
- Skiba, M.A.; Sikkema, A.P.; Moss, N.A.; Lowell, A.N.; Su, M.; Sturgis, R.M.; Gerwick, L.; Gerwick, W.H.; Sherman, D.H.; Smith, J.L. Biosynthesis of t-butyl in apratoxin A: Functional analysis and architecture of a PKS loading module. ACS Chem. Biol. 2018, 13, 1640–1650. [Google Scholar] [CrossRef]
- Wadsworth, A.D.; Furkert, D.P.; Brimble, M.A. Total synthesis of the macrocyclic N-Methyl enamides palmyrolide A and 2S-sanctolide A. J. Org. Chem. 2014, 79, 11179–11193. [Google Scholar] [CrossRef]
- Tello-Aburto, R.; Johnson, E.M.; Valdez, C.K.; Maio, W.A. Asymmetric total synthesis and absolute stereochemistry of the neuroactive marine macrolide palmyrolide A. Org. Lett. 2012, 14, 2150–2153. [Google Scholar] [CrossRef] [Green Version]
- Tello-Aburto, R.; Newar, T.D.; Maio, W.A. Evolution of a protecting-group-free total synthesis: Studies en route to the neuroactive marine macrolide (-)-palmyrolide A. J. Org. Chem. 2012, 77, 6271–6289. [Google Scholar] [CrossRef]
- Lam, N.Y.S.; Muir, G.; Challa, V.R.; Britton, R.; Paterson, I. A counterintuitive stereochemical outcome from a chelation-controlled vinylmetal aldehyde addition leads to the configurational reassignment of phormidolide A. Chem. Commun. 2019, 55, 9717–9720. [Google Scholar] [CrossRef]
- Helfrich, E.J.N.; Ueoka, R.; Dolev, A.; Rust, M.; Meoded, R.A.; Bhushan, A.; Califano, G.; Costa, R.; Gugger, M.; Steinbeck, C.; et al. Automated structure prediction of trans-acyltransferase polyketide synthase products. Nat. Chem. Biol. 2019, 15, 813–821. [Google Scholar] [CrossRef]
- Ndukwe, I.E.; Wang, X.; Lam, N.Y.S.; Ermanis, K.; Alexander, K.L.; Bertin, M.J.; Martin, G.E.; Muir, G.; Paterson, I.; Britton, R.; et al. Synergism of anisotropic and computational NMR methods reveals the likely configuration of phormidolide A. Chem. Commun. 2020, 56, 7565–7568. [Google Scholar] [CrossRef]
- Shinomiya, S.; Iwasaki, A.; Ohno, O.; Suenaga, K. Total synthesis and stereochemical determination of yoshinone A. Phytochemistry 2016, 132, 109–114. [Google Scholar] [CrossRef]
- Koyama, T.; Kawazoe, Y.; Iwasaki, A.; Ohno, O.; Suenaga, K.; Uemura, D. Anti-obesity activities of the yoshinone A and the related marine γ-pyrone compounds. J. Antibiot. 2016, 69, 348–351. [Google Scholar] [CrossRef]
- Mohamed, Z. Harmful cyanobacteria and their cyanotoxins in Egyptian fresh waters–state of knowledge and research needs. Afr. J. Aquat. Sci. 2016, 41, 361–368. [Google Scholar] [CrossRef]
- Joshi, D.; Mohandass, C.; Dhale, M. Effect of UV-B radiation and desiccation stress on photoprotective compounds accumulation in marine Leptolyngbya sp. Appl. Biochem. Biotechnol. 2018, 184, 35–47. [Google Scholar] [CrossRef]
- Babu, S.V.; Ashokkumar, B.; Sivakumar, N.; Sudhakarsamy, P.; Varalakshmi, P. Indole-3-acetic acid from filamentous cyanobacteria: Screening, strain identification and production. J. Sci. Ind. Res. 2013, 72, 581–584. [Google Scholar]
- Stevenson, C.S.; Capper, E.A.; Roshak, A.K.; Marquez, B.; Eichman, C.; Jackson, J.R.; Mattern, M.; Gerwick, W.H.; Jacobs, R.S.; Marshall, L.A. The identification and characterization of the marine natural product scytonemin as a novel antiproliferative pharmacophore. J. Pharmacol. Exp. Ther. 2002, 303, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Soule, T.; Stout, V.; Swingley, W.D.; Meeks, J.C.; Garcia-Pichel, F. Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. J. Bacteriol. 2007, 189, 4465–4472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balskus, E.P.; Walsh, C.T. Investigating the initial steps in the biosynthesis of cyanobacterial sunscreen scytonemin. J. Am. Chem. Soc. 2008, 130, 15260–15261. [Google Scholar] [CrossRef] [Green Version]
- Sorrels, C.M.; Proteau, P.J.; Gerwick, W.H. Organization, evolution, and expression analysis of the biosynthetic gene cluster for scytonemin, a cyanobacterial UV-absorbing pigment. Appl. Environ. Microbiol. 2009, 75, 4861–4869. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.S.; Esquenazi, E.; Dorrestein, P.C.; Gerwick, W.H. Probing the in vivo biosynthesis of scytonemin, a cyanobacterial ultraviolet radiation sunscreen, through small scale stable isotope incubation studies and MALDI-TOF mass spectrometry. Bioorg. Med. Chem. 2011, 19, 6620–6627. [Google Scholar] [CrossRef] [Green Version]
- Martins, S.; Mussatto, S.I.; Martínez-Avila, G.; Montañez-Saenz, J.; Aguilar, C.N.; Teixeira, J.A. Bioactive phenolic compounds: Production and extraction by solid-state fermentation. A review. Biotechnol. Adv. 2011, 29, 365–373. [Google Scholar] [CrossRef] [Green Version]
- Ljaz, S.; Hasnain, S. Antioxidant potential of indigenous cyanobacterial strains in relation with their phenolic and flavonoid contents. Nat. Prod. Res. 2016, 30, 1297–1300. [Google Scholar]
- Trabelsi, L.; Mnari, A.; Abdel-Daim, M.M.; Abid-Essafi, S.; Aleya, L. Therapeutic properties in Tunisian hot springs: First evidence of phenolic compounds in the cyanobacterium Leptolyngbya sp. biomass, capsular polysaccharides and releasing polysaccharides. BMC Complement. Altern. Med. 2016, 16, 515. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.L.; Boyer, G.L.; Zimba, P.V. A review of cyanobacterial odorous and bioactive metabolites: Impacts and management alternatives in aquaculture. Aquaculture 2008, 280, 5–20. [Google Scholar] [CrossRef]
- Wang, Z.; Xiao, P.; Song, G.; Li, Y.; Li, R. Isolation and characterization of a new reported cyanobacterium Leptolyngbya bijugata coproducing odorous geosmin and 2-methylisoborneol. Environ. Sci. Pollut. Res. 2015, 22, 12133–12140. [Google Scholar] [CrossRef] [PubMed]
- Schipper, K.; Fortunati, F.; Oostlander, P.C.; Al Muraikhi, M.; Al Jabri, H.M.S.J.; Wijffels, R.H.; Barbosa, M.J. Production of phycocyanin by Leptolyngbya sp. in desert environments. Algal Res. 2020, 47, 101875. [Google Scholar] [CrossRef]
- Singh, N.K.; Sonani, R.R.; Awasthi, A.; Prasad, B.; Patel, A.R.; Kumar, J.; Madamwar, D. Phycocyanin moderates aging and proteotoxicity in caenorhabditis elegans. J. Appl. Phycol. 2016, 28, 2407–2417. [Google Scholar] [CrossRef]
- Kim, S.-K. Handbook of Anticancer Drugs from Marine Origin; Springer: Berlin/Heidelberg, Germany, 2015; ISBN 9783319071442. [Google Scholar]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef]
- Videau, P.; Wells, K.N.; Singh, A.J.; Eiting, J.; Proteau, P.J.; Philmus, B. Expanding the natural products heterologous expression repertoire in the model cyanobacterium Anabaena sp. strain PCC 7120: Production of pendolmycin and teleocidin B-4. ACS Synth. Biol. 2020, 9, 63–75. [Google Scholar] [CrossRef]
- Taton, A.; Ecker, A.; Diaz, B.; Moss, N.A.; Anderson, B.; Leão, T.F.; Simkovsky, R.; Dorrestein, P.C.; Gerwick, L.; William, H. Heterologous expression of cryptomaldamide in a cyanobacterial host. BioRxiv 2020. [Google Scholar] [CrossRef]























| Name | Geographic Location | Culture | Total Synthesis | Bioactivity | Cell Line | Activity | Reference |
|---|---|---|---|---|---|---|---|
| Coibamide A (1) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | IC50 3.9 nM | [30] |
| Cytotoxicity | A549 | IC50 3.6 nM | |||||
| Cytotoxicity | MCF-7 | IC50 35.7 nM | |||||
| N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 5.0 nM | [31] | |
| Cytotoxicity | A549 | GI50 5.4 nM | |||||
| Cytotoxicity | PANC-1 | GI50 3.1 nM | |||||
| Coiba National Park, Panama | N a | N a | Cytotoxicity | H460 | LC50 < 23 nM | [32] | |
| Cytotoxicity | Mouse neuro2a | LC50 < 23 nM | |||||
| Cytotoxicity | MDA-MB-231 | GI50 2.8 nM | |||||
| Cytotoxicity | LOX IMVI | GI50 7.4 nM | |||||
| Cytotoxicity | HL-60(TB) | GI50 7.4 nM | |||||
| Cytotoxicity | SNB-75 | GI50 7.6 nM | |||||
| Histological Selectivity | Breast, CNS, colon, and ovarian cancer cells | Good | |||||
| Coiba National Park, Panama | N a | N a | Cytotoxicity | U87-MG | EC50 28.8 nM | [33] | |
| Cytotoxicity | SF-295 glioblastoma cell | EC50 96.2 nM | |||||
| Cytotoxicity | MEFs | EC50 96.2 nM | |||||
| Synthetic l-HVA, l-MeAla-Coibamide A (2) | N a | N a | Y b | Cytotoxicity | COLO205 | IC50 11.5 µM | [33] |
| Cytotoxicity | H460 | 45% inhibition at 20 µM | |||||
| Cytotoxicity | MDA-MB-231 | IC50 17.98 µM | [34] | ||||
| Cytotoxicity | MCF-7 | IC50 11.77 µM | |||||
| Cytotoxicity | A549 | IC50 22.80 µM | |||||
| Cytotoxicity | MDA-MB-231 | GI50 > 16000 nM | [31] | ||||
| Cytotoxicity | A549 | GI50 22800 nM | |||||
| Cytotoxicity | PANC-1 | ND c | |||||
| N a | N a | N a | Cytotoxicity | H292 | IC50 124 nM | [35] | |
| Cytotoxicity | MDA-MB-231 | IC50 66 nM | |||||
| Cytotoxicity | PC-3 | IC50 80 nM | |||||
| Cytotoxicity | SF-295 | IC50 219 nM | |||||
| Synthetic O-Desmethyl, l-HVA, l-MeAla-Coibamide A (3) | N a | N a | Y b | Cytotoxicity | COLO205 | IC50 13 µM | [33] |
| Cytotoxicity | H460 | 36% inhibition at 20 µM | |||||
| Synthetic l-HVA, d-MeAla-Coibamide A (4) | N a | N a | Y b | Cytotoxicity | A549 | IC50 19.0 nM | [35] |
| Cytotoxicity | HCT116 | IC50 44.6 nM | |||||
| Cytotoxicity | MCF-7 | IC50 48.6 nM | |||||
| Cytotoxicity | B16 | IC50 54.4 nM | |||||
| Cytotoxicity | H292 | IC50 610 nM | |||||
| Cytotoxicity | MDA-MB-231 | IC50 545 nM | |||||
| Cytotoxicity | PC-3 | IC50 424 nM | |||||
| Cytotoxicity | SF-295 | IC50 816 nM | |||||
| Cytotoxicity | MDA-MB-231 | GI50 545 nM | [31] | ||||
| Cytotoxicity | A549 | GI50 19 nM | |||||
| Cytotoxicity | PANC-1 | ND c | |||||
| Synthetic Coibamide A-1c (5) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 7518 nM | [31] |
| Cytotoxicity | A549 | GI50 20091 nM | |||||
| Cytotoxicity | PANC-1 | GI50 12417 nM | |||||
| Synthetic Coibamide A-1d (6) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 10809 nM | [31] |
| Cytotoxicity | A549 | ND c | |||||
| Cytotoxicity | PANC-1 | ND c | |||||
| Synthetic Coibamide A-1e (7) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 2662 nM | [31] |
| Cytotoxicity | A549 | GI50 1995 nM | |||||
| Cytotoxicity | PANC-1 | GI50 1906 nM | |||||
| Synthetic MeAla3-MeAla6-Coibamide A-1f (8) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 5.1 nM | [31] |
| Cytotoxicity | A549 | GI50 7.3 nM | |||||
| Cytotoxicity | PANC-1 | GI50 7.0 nM | |||||
| Synthetic Coibamide A-1g (9) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 5.3 nM | [31] |
| Cytotoxicity | A549 | GI50 12.4 nM | |||||
| Cytotoxicity | PANC-1 | GI50 32.9 nM | |||||
| Synthetic Coibamide A-1h (10) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 61.6 nM | [31] |
| Cytotoxicity | A549 | GI50 81.7 nM | |||||
| Cytotoxicity | PANC-1 | GI50 124 nM | |||||
| Synthetic Coibamide A-1i (11) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 20.8 nM | [31] |
| Cytotoxicity | A549 | GI50 194 nM | |||||
| Cytotoxicity | PANC-1 | GI50 46.3 nM | |||||
| Synthetic Coibamide A-1j (12) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 2056 nM | [31] |
| Cytotoxicity | A549 | ND c | |||||
| Cytotoxicity | PANC-1 | GI50 2178 nM | |||||
| Synthetic Coibamide A-1k (13) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 183 nM | [31] |
| Cytotoxicity | A549 | GI50 222 nM | |||||
| Cytotoxicity | PANC-1 | GI50 277 nM | |||||
| Synthetic Coibamide A-1l (14) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 450 nM | [31] |
| Cytotoxicity | A549 | GI50 473 nM | |||||
| Cytotoxicity | PANC-1 | GI50 601 nM | |||||
| Synthetic Coibamide A-1m (15) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 415 nM | [31] |
| Cytotoxicity | A549 | GI50 511 nM | |||||
| Cytotoxicity | PANC-1 | GI50 723 nM | |||||
| Synthetic Coibamide A-1n (16) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 >16000nM | [31] |
| Cytotoxicity | A549 | ND c | |||||
| Cytotoxicity | PANC-1 | ND c | |||||
| Synthetic Coibamide A-1o (17) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 470 nM | [31] |
| Cytotoxicity | A549 | GI50 733 nM | |||||
| Cytotoxicity | PANC-1 | GI50 828 nM | |||||
| Synthetic Coibamide A-1p (18) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 236 nM | [31] |
| Cytotoxicity | A549 | GI50 360 nM | |||||
| Cytotoxicity | PANC-1 | GI50 204 nM | |||||
| Synthetic Coibamide A-1q (19) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 239 nM | [31] |
| Cytotoxicity | A549 | GI50 443 nM | |||||
| Cytotoxicity | PANC-1 | GI50 415 nM | |||||
| Synthetic Coibamide A-1r (20) | N a | N a | Y b | Cytotoxicity | MDA-MB-231 | GI50 >16000 nM | [31] |
| Cytotoxicity | A549 | ND c | |||||
| Cytotoxicity | PANC-1 | ND c | |||||
| Dolastatin 12 (21) | The Red Sea | Y b | N a | Cytotoxicity | HeLa cells | IC50 > 1 μM | [36] |
| KB (human nasopharyngeal carcinoma cell line) | MICs <0.05 µg/mL | [37] | |||||
| LoVo (a human colon adenocarcinoma cell line) | 0.08 µg/mL | ||||||
| Murine P388 lymphocytic leukemia | ED50 7.5 × 10−2 µg/mL | [38,39] | |||||
| Ibu-Epidemethoxylyngbyastatin 3 (22) | The Red Sea | Y b | N a | Cytotoxicity | HeLa cells | IC50 > 10 μM | [36] |
| Grassypeptolide D (23) | The Red Sea | Y b | N a | Cytotoxicity | HeLa cells | IC50 335 nM | [36] |
| Cytotoxicity | Mouse neuro2a blastoma cells | IC50 599 nM | |||||
| Grassypeptolide E (24) | Cytotoxicity | HeLa cells | IC50 192 nM | [36] | |||
| Cytotoxicity | Mouse neuro2a blastoma cells | IC50 407 nM | |||||
| Loggerpeptin A (25) | Florida, USA | N a | N a | Antiproteolytic Activity | Bovine pancreatic chymotrypsin | IC50 0.24 μM | [40] |
| Porcine pancreatic elastase | IC50 0.24 μM | ||||||
| Human neutrophil elastase | IC50 0.29 μM | ||||||
| Loggerpeptin B (26) | Florida, USA | N a | N a | Antiproteolytic Activity | Bovine pancreatic chymotrypsin | IC50 0.22 μM | [40] |
| Porcine pancreatic elastase | IC50 0.28 μM | ||||||
| Human neutrophil elastase | IC50 0.89 μM | ||||||
| Loggerpeptin C (27) | Florida, USA | N a | N a | Antiproteolytic Activity | Bovine pancreatic chymotrypsin | IC50 0.35 μM | [40] |
| Porcine pancreatic elastase | IC50 0.54 μM | ||||||
| Human neutrophil elastase | IC50 0.62 μM | ||||||
| Molassamide (28) | Florida, USA | N a | N a | Antiproteolytic Activity | Bovine pancreatic chymotrypsin | IC50 0.24 μM | [40] |
| Porcine pancreatic elastase | IC50 0.05 μM | ||||||
| Human neutrophil elastase | IC50 0.11 μM | ||||||
| 2-Hydroxyethyl-11-Hydroxyhexadec-9-Enoate (29) | Gulf of Thailand | Y b | N a | Antibacterial Activities | Vibrio harveyi | MIC 250–1000 µg/mL | [41] |
| Antibacterial Activities | Vibrio parahaemolyticus | MIC 350–1000 µg/mL | |||||
| Honaucin A (30) | Hawaii, USA | N a | N a | Anti-Inflammatory Activity | LPS-stimulated RAW264.7 murine macrophages | IC50 4.0 µM | [42] |
| Antioxidant Activity | Radical Oxygen Scavenging | No activity at 146 µM | |||||
| QS-Inhibitory activities | V. harveyi BB120 | IC50 5.6 µM | |||||
| QS-Inhibitory activities | E. coli JB525 | IC50 38.5 µM | |||||
| Cytotoxicity | RAW264.7 cells | No activity at 1 µg/mL | [43] | ||||
| Cellular TRAP Activity | RANKL-induced osteoclastogenesis in RAW264.7 cells | IC50 0.63 μg/mL | |||||
| Honaucin B (31) | Hawaii, USA | N a | N a | Anti-Inflammatory Activity | LPS-stimulated RAW264.7 murine macrophages | IC50 4.5 µM | [42] |
| QS-Inhibitory activities | V. harveyi BB120 | IC50 17.6 µM | |||||
| QS-Inhibitory activities | E. coli JB525 | IC50 > 500 µM | |||||
| Honaucin C (32) | Hawaii, USA | N a | N a | Anti-Inflammatory Activity | LPS-stimulated RAW264.7 murine macrophages | IC50 7.8 µM | [42] |
| QS-Inhibitory activities | V. harveyi BB120 | IC50 14.6 µM | |||||
| QS-Inhibitory activities | E. coli JB525 | IC50 > 500 µM | |||||
| Synthetic Br-Honaucin A (33) | N a | N a | Y b | Cytotoxicity | RAW264.7 cells | No activity at 1 µg/mL | [43] |
| Cellular TRAP Activity | RANKL-induced osteoclastogenesis in RAW264.7 cells | IC50 0.54 μg/mL | |||||
| Synthetic Hex-Honaucin A (34) | N a | N a | Y b | Cytotoxicity | RAW264.7 cells | 71.6% cell viability at 1 µg/mL | [43] |
| Cellular TRAP Activity | RANKL-induced osteoclastogenesis in RAW264.7 cells | IC50 0.68 μg/mL | |||||
| Synthetic I-Honaucin A (35) | N a | N a | Y b | Cytotoxicity | RAW264.7 cells | No activity at 1 µg/mL | [43] |
| Cellular TRAP Activity | RANKL-induced osteoclastogenesis in RAW264.7 cells | IC50 0.61 μg/mL | |||||
| Leptolyngbyolide A (36) | Okinawa, Japan | N a | Y b | Cytotoxicity | HeLa S3 cell | IC50 0.099 µM | [44] |
| Actin-Depolymerizing activity | F-actin | EC50 12.6 µM | |||||
| Leptolyngbyolide B (37) | Okinawa, Japan | N a | Y b | Cytotoxicity | HeLa S3 cell | IC50 0.16 µM | [44] |
| Actin-Depolymerizing activity | F-actin | EC50 11.6 µM | |||||
| Leptolyngbyolide C (38) | Okinawa, Japan | N a | Y b | Cytotoxicity | HeLa S3 cell | IC50 0.64 µM | [44] |
| Actin-Depolymerizing activity | F-actin | EC50 26.9 µM | |||||
| Leptolyngbyolide D (39) | Okinawa, Japan | N a | Y b | Cytotoxicity | HeLa S3 cell | IC50 0.15 µM | [44] |
| Actin-Depolymerizing activity | F-actin | EC50 21.5 µM | |||||
| Palmyrolide A (40) | Palmyra Atoll | N a | Y b | Ca2+ Influx (Inhibition) | Murine cerebrocortical neurons | IC50 3.70 µM (2.29–5.98 µM, 95% CI) | [45] |
| Na+ Channel Blocking Activity | Mouse neuroblastoma (neuro2a) | IC50 5.2 µM | |||||
| Cytotoxicity | H460 | No activity at 20 µM | |||||
| Phormidolide (42) | The Red Sea | Y b | N a | Brine Shrimp Toxicity | LC50 1.5 µM | [46] | |
| Kalkipyrone A (43) | America Samoa | N a | N a | Cytotoxicity | H460 cells | EC50 0.9 µM | [47] |
| Cytotoxicity | Saccharomyces cerevisiae ABC16-monster | IC50 14.6 µM | |||||
| Brine Shrimp Toxicity | Brine shrimp (Artemia salina) | LD50 1 µg/mL | [48] | ||||
| Ichthyotoxicity | Goldfish Carassius auratus | LD50 2 µg/mL | |||||
| Cytotoxicity | NCI’s 60 human tumor cell line | Modestly inhibitory to several renal and melanoma cell lines | |||||
| Kalkipyrone B (44) | America Samoa | N a | N a | Cytotoxicity | H460 cells | EC50 9.0 µM | [47] |
| Cytotoxicity | Saccharomyces cerevisiae ABC16-monster | IC50 13.4 µM | |||||
| Yoshinone A (45) | Ishigaki island, Japan | N a | Y b | Adipogenic Differentiation | 3T3-L1 cells | EC50 420 nM | [49] |
| Cytotoxicity | 3T3-L1 cells | IC50 > 50 µM | |||||
| Cytotoxicity | HeLa | IC50 > 50 µM | |||||
| Cytotoxicity | Saccharomyces cerevisiae ABC16-monster | IC50 63.8 µM | [47] | ||||
| Cytotoxicity | H460 cells | EC50 > 10 µM | |||||
| Yoshinone B1 (46) | Ishigaki island, Japan | N a | N a | Adipogenic Differentiation | 3T3-L1 cells | <50% inhibition at 5 µM | [49] |
| Yoshinone B2 (47) | Ishigaki island, Japan | N a | N a | Adipogenic Differentiation | 3T3-L1 cells | <50% inhibition at 5 µM | [49] |
| Crossbyanol A (48) | Hawaii, USA | N a | N a | Cytotoxicity | H460 human lung cancer cells | IC50 30 µg/ mL | [50] |
| Na+ Influx (Activation and Inhibition) | Mouse neuroblastoma (neuro2a) | IC50 20 µg/mL(Activation) | |||||
| Antibacterial Activity | Methicillin-resistant Staphylococcus aureus (MRSA) | No activity at 125 µg/mL | |||||
| Brine Shrimp Toxicity | Brine shrimp (Artemia salina) | No activity at 25 µg/mL | |||||
| Crossbyanol B (49) | Hawaii, USA | N a | N a | Cytotoxicity | H460 human lung cancer cells | No activity at 20 µg/mL | [50] |
| Na+ Influx (Activation and Inhibition) | Mouse neuroblastoma (neuro2a) | No activity at 20 µg/mL | |||||
| Antibacterial activity | Methicillin-resistant Staphylococcus aureus (MRSA) | MIC 2.0–3.9 µg/mL | |||||
| Brine Shrimp Toxicity | Brine shrimp (Artemia salina) | IC50 2.8 µg/mL | |||||
| Crossbyanol C (50) | Hawaii, USA | N a | N a | Cytotoxicity | H460 human lung cancer cells | No activity at 20 µg/mL | [50] |
| Na+ Influx (Activation and Inhibition) | Mouse neuroblastoma (neuro2a) | No activity at 20 µg/mL | |||||
| Crossbyanol D (51) | Hawaii, USA | N a | N a | Cytotoxicity | H460 human lung cancer cells | No activity at 20 µg/mL | [50] |
| Na+ Influx (Activation and Inhibition) | Mouse neuroblastoma (neuro2a) | No activity at 20 µg/mL | |||||
| Leptazoline A (52) | Honolulu | Y b | N a | Cytotoxicity | PANC-1 | No significant activity | [51] |
| Leptazoline B (53) | Honolulu | Y b | N a | Cytotoxicity | PANC-1 | GI50 10 µM |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Naman, C.B.; Alexander, K.L.; Guan, H.; Gerwick, W.H. The Chemistry, Biochemistry and Pharmacology of Marine Natural Products from Leptolyngbya, a Chemically Endowed Genus of Cyanobacteria. Mar. Drugs 2020, 18, 508. https://doi.org/10.3390/md18100508
Li Y, Naman CB, Alexander KL, Guan H, Gerwick WH. The Chemistry, Biochemistry and Pharmacology of Marine Natural Products from Leptolyngbya, a Chemically Endowed Genus of Cyanobacteria. Marine Drugs. 2020; 18(10):508. https://doi.org/10.3390/md18100508
Chicago/Turabian StyleLi, Yueying, C. Benjamin Naman, Kelsey L. Alexander, Huashi Guan, and William H. Gerwick. 2020. "The Chemistry, Biochemistry and Pharmacology of Marine Natural Products from Leptolyngbya, a Chemically Endowed Genus of Cyanobacteria" Marine Drugs 18, no. 10: 508. https://doi.org/10.3390/md18100508