NIR Instruments and Prediction Methods for Rapid Access to Grain Protein Content in Multiple Cereals
Abstract
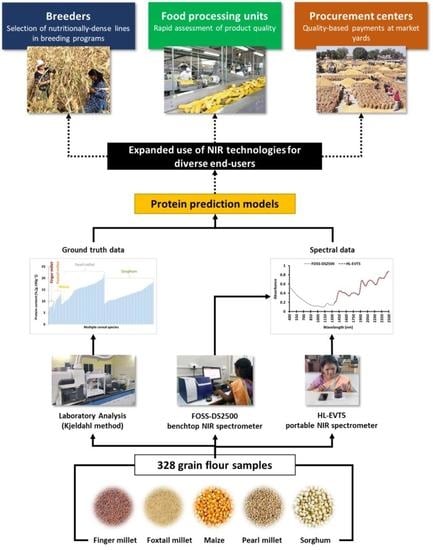
:1. Introduction
- (i)
- Compare the NIR spectra produced by benchtop (DS2500, FOSS) and portable (HL-EVT5, Hone) instruments, and their suitability for predicting grain protein in multiple cereal species;
- (ii)
- Assess the suitability of different model-building methods (via winISI, FOSS; Hone Create, Hone; and a customized CNN-based method) to predict protein content in multiple cereals using two types of NIR spectra;
- (iii)
- Ascertain the predictions made using multiple instrument–method combinations (i.e., FOSS-DS2500–winISI; FOSS-DS2500–Hone Create; FOSS-DS2500–CNN; HL-EVT5–Hone Create; and HL-EVT5–CNN) and discuss the suitability of their applications (for example, in decentralized breeding programs and markets).
2. Materials and Methods
2.1. Plant Material
2.2. Sample Collection and Preparation
2.3. Laboratory Analysis of Grain Protein Content (“Ground Truth” Dataset)
2.4. Scanning Samples with Two NIR Instruments
2.5. Calibration Model Development
2.5.1. Definition of Calibration and Validation Datasets
2.5.2. Prediction Method Development Using Established Software and a Custom-Made Pipeline: Instrument–Method Combinations
- FOSS-DS2500 NIR Spectra Processed using WinISI Software:
- FOSS-DS2500 and HL-EVT5 NIR Spectra Processed using Hone Create Software:
- FOSS-DS2500 and HL-EVT5 NIR Spectra Processed using a Customized CNN-Based Algorithm:
2.6. Prediction Method Evaluation
3. Results
3.1. Diversity of Grain Protein Content in Five Cereal Species
3.2. NIR Spectrum Obtained from the Benchtop FOSS-DS2500 and the Portable HL-EVT5
3.3. NIR Spectrum Generated Using the FOSS-DS2500 and Processed via WinISI Software
3.4. NIR Spectrum Generated Using the FOSS-DS2500 and HL-EVT5, Processed via Hone Create Software
3.5. NIR Spectrum Generated Using FOSS-DS2500 and HL-EVT5, Processed via CNN-Based Customized Pipeline
3.6. Instrument–Method Combination Comparisons for Protein Content Predictions in Cereal Grains
4. Discussion
4.1. Importance of NIR Spectroscopy for Rapid Cereal Grain Quality Assessment
4.2. Expected Data Properties as Prerequisites to Building Reliable Prediction Models
4.3. Prediction Methods for the Estimation of Protein Content in Multiple Cereal Grains and Their Accuracy Metrics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agelet, L.E.; Hurburgh, C.R., Jr. A tutorial on near infrared spectroscopy and its calibration. Crit. Rev. Anal. Chem. 2010, 40, 246–260. [Google Scholar] [CrossRef]
- Workman, J.; Weyer, L. Practical Guide and Spectral Atlas for Interpretive Near-Infrared Spectroscopy, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar] [CrossRef]
- Villamuelas, M.; Serrano, E.; Espunyes, J.; Fernández, N.; López-Olvera, J.R.; Garel, M.; Santos, J.; Parra-Aguado, M.Á.; Ramanzin, M.; Fernández-Aguilar, X.; et al. Predicting herbivore faecal nitrogen using a multispecies near-infrared reflectance spectroscopy calibration. PLoS ONE 2017, 12, e0176635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rukundo, I.R.; Danao, M.G.C.; Mitchell, R.B.; Masterson, S.D.; Weller, C.L. Comparing the use of portable and benchtop NIR spectrometers in predicting nutritional value of forage. Appl. Eng. Agric. 2021, 37, 171–181. [Google Scholar] [CrossRef]
- FOSS-DS2500 Flour Analyzer from FOSS. Available online: https://www.dksh.com/global-en/products/ins/foss-flour-analyzer-nirs-ds2500 (accessed on 7 January 2021).
- Bruker-Tango FT-NIR Spectrometer from Bruker. Available online: https://www.bruker.com/en/products-and-solutions/infrared-and-raman/ft-nir-spectrometers/tango-ft-nir-spectrometer.html (accessed on 7 January 2021).
- Perten-IM9520 Flour Analyzer from PerkinElmer. Available online: https://www.calibrecontrol.com/main-product-list/perten-im9520-flour-analyser (accessed on 7 January 2021).
- Sorak, D.; Herberholz, L.; Iwascek, S.; Altinpinar, S.; Pfeifer, F.; Siesler, H.W. New developments and applications of portable raman, mid-infrared, and near-infrared spectrometers. Appl. Spectrosc. Rev. 2012, 47, 83–115. [Google Scholar] [CrossRef]
- Sarikaş, A.; Başar, M.D. An electronic portable device design to spectroscopically assess fruit quality. Turk. J. Electr. Eng. Comput. Sci. 2017, 25, 4063–4076. [Google Scholar] [CrossRef]
- Crocombe, R.A. Portable Spectroscopy. Appl. Spectrosc. 2018, 72, 1701–1751. [Google Scholar] [CrossRef]
- MicroNIR OnSite-W from VIAVI Solutions. Available online: https://www.viavisolutions.com/en-us/osp/products/micronir-onsite-w (accessed on 7 January 2021).
- DLP NIRScanTM Nano EVM Spectrometer from Texas Instruments. Available online: https://www.ti.com/tool/DLPNIRNANOEVM (accessed on 7 January 2021).
- MEMS Spectrometer from Fraunhofer. Available online: https://www.ipms.fraunhofer.de/en/Components-and-Systems/Components-and-Systems-Sensors/Optical-Sensors/MEMS-based-spectroscopy.html (accessed on 7 January 2021).
- Hone Lab Red from Hone. Available online: https://www.honeag.com/hone-lab (accessed on 7 January 2021).
- Osborne, B.G. Near infrared spectroscopy in food analysis. In Encyclopedia of Analytical Chemistry: Applications, Theory and Instrumentation; John Wiley & Sons Ltd: New York, NY, USA, 2006. [Google Scholar] [CrossRef]
- Singh, C.B.; Paliwal, J.; Jayas, D.S.; White, N.D. Near-infrared spectroscopy: Applications in the grain industry. In Proceedings of the CSBE/SCGAB Annual Conference, Edmonton, Alberta, 16–19 July 2006. [Google Scholar]
- dos Santos, C.A.T.; Lopo, M.; Páscoa, R.N.M.J.; Lopes, J.A. A Review on the Applications of portable near-infrared spectrometers in the agro-food industry. Appl. Spectrosc. 2013, 67, 215–1233. [Google Scholar] [CrossRef]
- Williams, P.C. Application of near infrared reflectance spectroscopy to analysis of cereal grains and oilseeds. Cereal Chem. 1975, 52, 561–576. [Google Scholar]
- Norris, K.H.; Barnes, R.F.; Moore, J.E.; Shenk, J.S. Predicting forage quality by infrared reflectance spectroscopy. J. Anim. Sci. 1976, 43, 889–897. [Google Scholar] [CrossRef]
- Estienne, F.; Pasti, L.; Centner, V.; Walczak, B.; Despagne, F.; Rimbaud, D.J.; De Noord, O.E.; Massart, D.L. A comparison of multivariate calibration techniques applied to experimental NIR data sets: Part II. Predictive ability under extrapolation conditions. Chemometr. Intell. Lab. Syst. 2001, 58, 195–211. [Google Scholar] [CrossRef] [Green Version]
- Esbensen, K.H.; Julius, L.P. Representative sampling, data quality, validation—A necessary trinity in chemometrics. In Comprehensive Chemometrics; Brown, S., Tauler, R., Walczak, R., Eds.; Elsevier: Oxford, UK, 2009; Volume 4, pp. 1–20. [Google Scholar]
- Agelet, L.E.; Hurburgh, C.R., Jr. Limitations and current applications of near infrared spectroscopy for single seed analysis. Talanta 2014, 121, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Zhu, L.; Lou, X.; Meng, X.; Guo, Y.; Wang, Z. A new local modelling approach based on predicted errors for near-infrared spectral analysis. J. Anal. Methods Chem. 2016, 2016, 5416506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheewapramong, P. Use of Near-Infrared Spectroscopy for Qualitative and Quantitative Analyses of Grains and Cereal Products. Ph.D. Thesis, University of Nebraska-Lincoln, Lincoln, NE, USA, 2007. [Google Scholar]
- Downey, G. NIR and chemometrics in the service of the food industry. NIR News 2007, 18, 10–11. [Google Scholar] [CrossRef]
- Chen, J.Y.; Miao, Y.; Sato, S.; Zhang, H. Near infrared spectroscopy for determination of the protein composition of rice flour. Food Sci. Technol. Res. 2008, 14, 132–138. [Google Scholar] [CrossRef] [Green Version]
- Kahriman, F.; Egesel, C.Ö. Development of a calibration model to estimate quality traits in wheat flour using NIR (Near Infrared Reflectance) spectroscopy. Res. J. Agric. Sci. 2011, 43, 392–400. [Google Scholar]
- Bagchi, T.B.; Sharma, S.; Chattopadhyay, K. Development of NIRS models to predict protein and amylose content of brown rice and proximate compositions of rice bran. Food Chem. 2016, 191, 21–27. [Google Scholar] [CrossRef]
- Lyu, N.; Chen, J.; Pan, T.; Yao, L.; Han, Y.; Yu, J. Near-infrared spectroscopy combined with equidistant combination partial least squares applied to multi-index analysis of corn. Infrared Phys. Techn. 2016, 76, 648–654. [Google Scholar] [CrossRef]
- Sampaio, P.S.; Soares, A.; Castanho, A.; Almeida, A.S.; Oliveira, J.; Brites, C. Optimization of rice amylose determination by NIR-spectroscopy using PLS chemometrics algorithms. Food Chem. 2018, 242, 196–204. [Google Scholar] [CrossRef]
- Tomas, E.; Bayram, I. Establishing near infrared spectroscopy (NIR) calibration for starch analysis in corn grain. Kocatepe Vet. J. 2018, 12, 7–14. [Google Scholar]
- Chen, J.; Li, M.; Pan, T.; Pang, L.; Yao, L.; Zhang, J. Rapid and non-destructive analysis for the identification of multi-grain rice seeds with near-infrared spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 219, 179–185. [Google Scholar] [CrossRef]
- Kahriman, F.; Liland, K.H. SelectWave: A graphical user interface for wavelength selection and spectral data analysis. Chemom. Intell. Lab. Syst. 2021, 212, 104275. [Google Scholar] [CrossRef]
- Lee, S.; Choi, H.; Cha, K.; Kim, M.-K.; Kim, J.-S.; Youn, C.H.; Lee, S.-H.; Chung, H. Random Forest as a non-parametric algorithm for near-infrared (NIR) spectroscopic discrimination for geographical origin of agricultural samples. Bull. Korean Chem. Soc. 2012, 33, 4267–4270. [Google Scholar] [CrossRef] [Green Version]
- Kong, W.; Zhang, C.; Liu, F.; Nie, P.; He, Y. Rice seed cultivar identifcation using near-infrared hyperspectral imaging and multivariate data analysis. Sensors 2013, 13, 8916–8927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Tan, C.; Lin, Z. Authenticity detection of black rice by near-infrared spectroscopy and support vector data description. Int. J. Anal. Chem. 2018, 2018, 8032831. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Fearn, T. Modern practical convolutional neural networks for multivariate regression: Applications to NIR calibration. Chemometr Intell. Lab. Syst. 2018, 182, 9–20. [Google Scholar] [CrossRef]
- Das, B.; Nair, B.; Reddy, V.K.; Venkatesh, P. Evaluation of multiple linear, neural network and penalised regression models for prediction of rice yield based on weather parameters for west coast of India. Int. J. Biometeorol. 2018, 62, 1809–1822. [Google Scholar] [CrossRef]
- Le, T.H.; Chen, H.; Babar, M.A. Deep learning for source code modeling and generation: Models, applications, and challenges. ACM Comput. Surv. 2020, 53, 62–100. [Google Scholar] [CrossRef]
- Sampaio, P.S.; Castanho, A.; Almeida, A.S.; Oliveira, J.; Brites, C. Identification of rice flour types with near-infrared spectroscopy associated with PLS-DA and SVM methods. Eur. Food Res. Technol. 2020, 246, 527–537. [Google Scholar] [CrossRef]
- Kabir, M.H.; Guindo, M.L.; Chen, R.; Liu, F. Geographic origin discrimination of millet using vis-NIR spectroscopy combined with machine learning techniques. Foods 2021, 11, 2767. [Google Scholar] [CrossRef]
- Kahriman, F.; Egesel, C.Ö. Using near infrared (NIR) spectroscopy in the analysis of cereal products: The example of maize. In Recent Researches in Science and Landscape Management; Efe, R., Zencirkiran, M., Curebal, İ., Eds.; Cambridge Scholars Publishing: Newcastle, NSW, Australia, 2018; pp. 507–521. [Google Scholar]
- Ejaz, I.; He, S.; Li, W.; Hu, N.; Tang, C.; Li, S.; Li, M.; Diallo, B.; Xie, G.; Yu, K. Sorghum grains grading for food, feed, and fuel using NIR spectroscopy. Front. Plant Sci. 2021, 12, 720022. [Google Scholar] [CrossRef]
- Osborne, B.G.; Mertens, B.; Thompson, M.; Fearn, T. The authentication of Basmati rice using near infrared spectroscopy. J. Near Infrared Spectrosc. 1993, 1, 77–83. [Google Scholar] [CrossRef]
- Wang, H.L.; Wan, X.Y.; Bi, J.C.; Wang, J.K.; Jiang, L.; Chen, L.M.; Zhai, H.Q.; Wan, J.M. Quantitative analysis of fat content in rice by near-infrared spectroscopy technique. Cereal Chem. 2006, 83, 402–406. [Google Scholar] [CrossRef]
- Barnaby, J.Y.; Huggins, T.D.; Lee, H.; Mcclung, A.M.; Pinson, S.R.M.; Oh, M.; Bauchan, G.R.; Tarpley, L.; Lee, K.J.; Kim, M.S.; et al. Vis/NIR hyperspectral imaging production environment, and physicochemical grain properties in rice. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Burestan, F.N.; Sayyah, A.H.; Taghinezhad, E. Prediction of some quality properties of rice and its flour by near-infrared spectroscopy (NIRS) analysis. Food Sci. Nutr. 2020, 9, 1099–1105. [Google Scholar] [CrossRef]
- Fazeli, N.; Amir, B.; Afkari, H.; Mahdi, S. Prediction of amylose content, protein content, breakdown, and setback viscosity of Kadus rice and its flour by near-infrared spectroscopy (NIRS) analysis. J. Food Process. Preserv. 2020, 45, e15069. [Google Scholar]
- De Alencar Figueiredo, L.F.; Davrieux, F.; Fliedel, G.; Rami, J.F.; Chantereau, J.; Deu, M.; Courtois, B.; Mestres, C. Development of NIRS equations for food grain quality traits through exploitation of a core collection of cultivated sorghum. J. Agric. Food Chem. 2006, 54, 8501–8509. [Google Scholar] [CrossRef]
- Alfieri, M.; Cabassi, G.; Habyarimana, E.; Quaranta, F.; Balconi, C.; Redaelli, R. Discrimination and prediction of polyphenolic compounds and total antioxidant capacity in sorghum grains. JNIRS 2019, 27, 46–53. [Google Scholar] [CrossRef]
- Pojić, M.M.; Mastilović, J.S. Near infrared spectroscopy—advanced analytical tool in wheat breeding, trade, and processing. Food Bioprocess Technol. 2013, 6, 330–352. [Google Scholar] [CrossRef]
- Kahriman, F.; Egesel, C.Ö. Comparison of spectral and molecular analyses for classification of long term stored wheat samples. Guang Pu Xue Yu Guang Pu Fen Xi Guang Pu 2016, 36, 1266–1272. [Google Scholar]
- Levasseur-Garcia, C. Updated overview of infrared spectroscopy methods for detecting mycotoxins on cereals (Corn, Wheat, and Barley). Toxins 2018, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- Baeten, V.; Pierna, J.F.; Vermeulen, P.; Lecler, B.; Minet, O.; Zio, D.; Dardenne, P. Performance comparison of bench-top, hyperspectral imaging and pocket near infrared spectrometers: The example of protein quantification in wheat flour. In Proceedings of the 18th International Conference on Near Infrared Spectroscopy, Copenhagen, Denmark, 11–15 June 2017; Engelsen, S.B., Sørensen, K.M., van den Berg, F., Eds.; IM Publications Open: Chichester, UK, 2019; pp. 151–155. [Google Scholar]
- Johnson, J.B. An overview of near-infrared spectroscopy (NIRS) for the detection of insect pests in stored grains. J. Stored Prod. Res. 2020, 86, 101558. [Google Scholar] [CrossRef]
- Egesel, C.Ö.; Kahriman, F. Determination of quality parameters in maize grain by NIR reflectance spectroscopy. Tarim Bilim. Derg. 2012, 18, 31–42. [Google Scholar] [CrossRef]
- Egesel, C.Ö.; Kahriman, F.; Ekinci, N.; Kavdir, İ.; Büyükcan, M.B. Analysis of fatty acids in kernel, flour, and oil samples of maize by NIR spectroscopy using conventional regression. Cereal Chem. 2016, 93, 487–492. [Google Scholar] [CrossRef]
- Kahriman, F.; Onac, I.; Turk, F.; Öner, F.; Egesel, C.Ö. Determination of carotenoid and tocopherol content in maize flour and oil samples using near-infrared spectroscopy. Spectrosc. Lett. 2019, 52, 473–481. [Google Scholar] [CrossRef]
- Kahriman, F.; Onaç, I.; Oner, F.; Mert, F.; Egesel, C.Ö. Analysis of secondary biochemical components in maize flour samples by NIR (Near İnfrared Reflectance) Spectroscopy. J. Food Meas. Charact. 2020, 14, 2320–2332. [Google Scholar] [CrossRef]
- Serment, M.; Kahriman, F. Ability of near infrared spectroscopy and chemometrics to measure the phytic acid content in maize flour. Spectrosc. Lett. 2021, 54, 520–527. [Google Scholar] [CrossRef]
- Abeshu, Y. Developing Calibration Model for Prediction of Malt Barley and Teff Genotypes Quality Traits Using Near Infrared Spectroscopy (NIRS). Ph.D. Thesis, Addis Ababa University, Addis Ababa, Ethiopia, 2019. [Google Scholar]
- Abeshu, Y. Development of NIRS re-calibration model for ethiopian barley (Hordeum vulgare) lines traits to determine their brewing potential. J. Agric. Food Inf. 2021, 1, 100238. [Google Scholar] [CrossRef]
- Albanell, E.; Martínez, M.; De Marchi, M.; Manuelian, C.L. Prediction of bioactive compounds in barley by near-infrared reflectance spectroscopy (NIRS). J. Food Compos. Anal. 2021, 97, 103763. [Google Scholar] [CrossRef]
- Stubbs, T.L.; Kennedy, A.C.; Fortuna, A.M. Using NIRS to predict fiber and nutrient content of dryland cereal cultivars. J. Agric. Food Chem. 2010, 58, 398–403. [Google Scholar] [CrossRef]
- Rosales, A.; Galicia, L.; Oviedo, E.; Islas, C.; Palacios-Rojas, N. Near-infrared reflectance spectroscopy (NIRS) for protein, tryptophan, and lysine evaluation in quality protein maize (QPM) breeding programs. J. Agric. Food Chem. 2011, 59, 10781–10786. [Google Scholar] [CrossRef]
- Piaskowski, J.L.; Brown, D.; Campbell, K.G. Near-infrared calibration of soluble stem carbohydrates for predicting drought tolerance in spring wheat. Agron. J. 2016, 108, 285–293. [Google Scholar] [CrossRef] [Green Version]
- Norman, H.C.; Hulm, E.; Humphries, A.W.; Hughes, S.J.; Vercoe, P.E. Broad near-infrared spectroscopy calibrations can predict the nutritional value of >100 forage species within the Australian feedbase. Anim. Prod. Sci. 2020, 60, 1111–1122. [Google Scholar] [CrossRef]
- Zerihun, M.; Fox, G.; Nega, A.; Seyoum, A.; Minuye, M.; Jordan, D.; Taddese, T.; Assefa, A. Near-Infrared Reflectance Spectroscopy (NIRS) for Tannin, Starch and Amylase Determination in Sorghum Breeding Programs. J. Food Nutr. Sci. 2020, 7, 45–50. [Google Scholar] [CrossRef]
- Carreira, E.; Serrano, J.; Shahidian, S.; Nogales-Bueno, J.; Rato, A.E. Real-time quantification of crude protein and neutral detergent fibre in pastures under montado ecosystem using the portable NIR spectrometer. Appl. Sci. 2021, 11, 10638. [Google Scholar] [CrossRef]
- Smartfood-International Year of Millets. Available online: https://www.smartfood.org/international-year-of-millets-2023/millet (accessed on 8 February 2022).
- Sustainable Development Goal 3. Available online: https://in.one.un.org/page/sustainable-development-goals/sdg-3-2/ (accessed on 8 February 2022).
- Mainstreaming Millets. Available online: https://pib.gov.in/PressReleasePage.aspx?PRID=1783716 (accessed on 8 February 2022).
- Li, X.; Siddique, K.H. Future smart food: Harnessing the potential of neglected and underutilized species for Zero Hunger. Matern. Child Nutr. 2020, 16, 13008. [Google Scholar] [CrossRef] [PubMed]
- McKevith, B. Nutritional aspects of cereals. Nutr. Bull 2004, 29, 111–142. [Google Scholar] [CrossRef]
- Girish, C.; Meena, R.K.; Mahima, D.; Mamta, K. Nutritional properties of minor millets: Neglected cereals with potentials to combat malnutrition. Curr. Sci. 2014, 107, 1109–1111. [Google Scholar]
- Diao, X. Production and genetic improvement of minor cereals in China. Crop. J. 2017, 5, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Dodevska, M.S.; Djordjevic, B.I.; Sobajic, S.S.; Miletic, I.D.; Djordjevic, P.B.; Dimitrijevic-Sreckovic, V.S. Characterization of dietary fibre components in cereals and legumes used in Serbian diet. Food Chem. 2013, 141, 1624–1629. [Google Scholar] [CrossRef]
- Belesova, K.; Gasparrini, A.; Sié, A.; Sauerborn, R.; Wilkinson, P. Household cereal crop harvest and children’s nutritional status in rural Burkina Faso. Environ. Health 2017, 16, 1. [Google Scholar] [CrossRef] [Green Version]
- Rankoana, S.A. The use of indigenous knowledge in subsistence farming: Implications for sustainable agricultural production in dikgale community in Limpopo Province, South Africa. Towar. Sustain. Agric. Farming Pract. Water Use 2017, 63, 63–72. [Google Scholar]
- Yang, Z.; Han, L.; Li, Q. Discriminant analysis of meat and bone meal content in ruminant feed based on NIRS. Trans. Chin. Soc. Agric. Eng. 2009, 40, 124–128. [Google Scholar]
- García, J.; Cozzolino, D. Use of near infrared reflectance (NIR) spectroscopy to predict chemical composition of forages in broad-based calibration models. Agric. Téc. 2006, 66, 41–47. [Google Scholar] [CrossRef]
- Black, J.L.; Hughes, R.J.; Nielsen, S.G.; Tredrea, A.M.; Flinn, P.C. Near infrared reflectance analysis of grains to estimate nutritional value for chickens. In Proceedings of the 20th Australian Poultry Science Symposium, Sydney, NSW, Australia, 9–11 February 2009; Poultry Research Foundation: Brownlow Hill, NSW, Australia, 2009; pp. 31–34. [Google Scholar]
- Tahir, M.; Shim, M.Y.; Ward, N.E.; Smith, C.; Foster, E.; Guney, A.C.; Pesti, G.M. Phytate and other nutrient components of feed ingredients for poultry. Poult. Sci. 2012, 91, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Atalay, H.; Kahriman, F.; Alatürk, F. Estimation of dry matter, crude protein and starch values in mixed feeds by near-infrared reflectance (NIR). J. İst. Vet. Sci. 2020, 4, 125–130. [Google Scholar] [CrossRef]
- Atalay, H.; Kahrıman, F. Estimating roughage quality with near infrared reflectance (NIR) spectroscopy and chemometric techniques. Kocatepe Vet. J. 2020, 13, 234–240. [Google Scholar] [CrossRef]
- Kahriman, F.; Atalay, H. Estimation of relative feed value, relative forage quality and net energy lactation values of some roughage samples by using near infrared reflectance spectroscopy. J. Ist. Vet. Sci. 2020, 4, 109–118. [Google Scholar] [CrossRef]
- Cherney, J.H.; Digman, M.F.; Cherney, D.J. Portable NIRS for forage evaluation. Comput. Electron. Agric. 2021, 190, 106469. [Google Scholar] [CrossRef]
- Assadzadeh, S.; Walker, C.K.; McDonald, L.S.; Maharjan, P.; Panozzo, J.F. Multi-task deep learning of near infrared spectra for improved grain quality trait predictions. J. Near Infrared Spectrosc. 2020, 28, 275–286. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Kim, J.B.; Lee, S.Y.; Lee, H.S.; Gwag, J.G.; Kim, C.K.; Lee, Y.B. Application of near-infrared reflectance spectroscopy to rapid determination of seed fatty acids in foxtail millet (Setaria italica (L.) P. Beauv) germplasm. Korean J. Breed. Sci. 2010, 42, 448–454. [Google Scholar]
- Lee, Y.-Y.; Kim, J.-B.; Lee, H.-S.; Jeon, Y.-A.; Lee, S.-Y.; Kim, C.-K. Evaluation of millet (Panicum miliaceum subsp. miliaceum) germplasm for seed fatty acids using near-infrared reflectance spectroscopy. Korean J. Crop Sci. 2012, 57, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.-S.; Wang, L.-L.; Zhou, X.-R.; Shuang, S.-M.; Zhu, Z.-H.; Li, N.; Li, Y.; Liu, F.; Liu, S.-C.; Lu, P.; et al. Determination of protein, fat, starch, and amino acids in foxtail millet Setaria italica (L.) Beauv. by Fourier transform near-infrared reflectance spectroscopy. Food Sci. Biotechnol. 2013, 22, 1495–1500. [Google Scholar] [CrossRef]
- Bhardwaj, R.; Yadav, S.; Suneja, P. NIRS based food quality assessment approaches for cereals, oilseeds, pulses, fruits and vegetables. In Proceedings of the 7th Indo-Global Summit and Expo on Food & Beverages, New Delhi, India, 8–10 October 2015. [Google Scholar]
- Wheat Trading Standards in Australia. Available online: https://www.graintrade.org.au/commodity_standards (accessed on 8 February 2022).
- Wheat quality and Markets in Queensland, Department of Agriculture and Fisheries, Queensland. Available online: https://www.daf.qld.gov.au/__data/assets/pdf_file/0006/53799/Wheat-FactSheet-Quality-Markets-Qld.pdf (accessed on 8 February 2022).
- Huck, C.W. New Trend in Instrumentation of NIR Spectroscopy—Miniaturization. In Near-Infrared Spectroscopy; Ozaki, Y., Huck, C., Tsuchikawa, S., Engelsen, S.B., Eds.; Springer: Singapore, 2021; pp. 193–210. [Google Scholar] [CrossRef]
- Genebank of ICRISAT. Available online: https://www.genebank.icrisat.org (accessed on 22 December 2021).
- Upadhyaya, H.D.; Pundir, R.P.S.; Dwivedi, S.L.; Gowda, C.L.L.; Reddy, V.G.; Singh, S. Developing a mini core collection of sorghum for diversified utilization of germplasm. Crop Sci. 2009, 49, 1769–1780. [Google Scholar] [CrossRef] [Green Version]
- Jordan, D.R.; Mace, E.S.; Cruickshank, A.W.; Hunt, C.H.; Henzell, R.G. Exploring and exploiting genetic variation from unadapted sorghum germplasm in a breeding program. Crop Sci. 2011, 51, 1444–1457. [Google Scholar] [CrossRef]
- Deshpande, S.; Rakshit, S.; Manasa, K.G.; Pandey, S.; Gupta, R. Genomic Approaches for Abiotic Stress Tolerance in Sorghum. In The Sorghum Genome. Compendium of Plant Genomes; Springer: Berlin/Heidelberg, Germany, 2016; pp. 169–187. [Google Scholar]
- Kassahun, B.; Bidinger, F.R.; Hash, C.T.; Kuruvinashetti, M.S. Stay-green expression in early generation sorghum [Sorghum bicolor (L.) Moench] QTL introgression lines. Euphytica 2010, 172, 351–362. [Google Scholar] [CrossRef] [Green Version]
- Sehgal, D.; Skot, L.; Singh, R.; Srivastava, R.K.; Das, S.P.; Taunk, J.; Sharma, P.C.; Pal, R.; Raj, B.; Hash, C.T.; et al. Exploring potential of pearl millet germplasm association panel for association mapping of drought tolerance traits. PLoS ONE 2015, 10, e0122165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ICAR. Handbook of Agriculture. In Directorate of Publications and Information on Agriculture; ICAR Publication: New Delhi, India, 2011. [Google Scholar]
- Association of Official Analytical Chemists (AOAC) International. Official Methods of Analysis, 17th ed.; Association of Official Analytical Chemists (AOAC) International: Gaithersberg, MD, USA, 2000. [Google Scholar]
- Samireddypalle, A.; Boukar, O.; Grings, E.; Fatokun, C.A.; Kodukula, P.; Devulapalli, R.; Okike, I.; Blümmel, M. Cowpea and groundnut haulms fodder trading and its lessons for multidimensional cowpea improvement for mixed crop livestock systems in West Africa. Front. Plant Sci. 2017, 8, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayawardana, S.A.S.; Samarasekera, J.K.R.R.; Hettiarachchi, G.H.C.M.; Gooneratne, J.; Mazumdar, S.D.; Banerjee, R. Dietary fibers, starch fractions and nutritional composition of finger millet varieties cultivated in Sri Lanka. J. Food Compost. Anal. 2019, 82, 103249. [Google Scholar] [CrossRef] [Green Version]
- Hone Lab Video. Available online: https://www.youtube.com/watch?v=c7f_p3p-SVg (accessed on 8 February 2022).
- Hone Create Platform. Available online: https://www.honecreate.com (accessed on 8 February 2022).
- Williams, P. Calibration development and evaluation methods B. Set-up and evaluation. NIR News 2013, 24, 20–24. [Google Scholar] [CrossRef]
- Galvao, R.; Araujo, M.; Jose, G.; Pontes, M.; Silva, E.; Saldanha, T. A method for calibration and validation subset partitioning. Talanta 2005, 67, 736–740. [Google Scholar] [CrossRef]
- Kemps, B.J.; Saeys, W.; Mertens, K.; Darius, P.; De Baerdemaeker, J.G.; De Ketelaere, B. The importance of choosing the right validation strategy in inverse modelling. JNIRS 2010, 18, 231–237. [Google Scholar] [CrossRef]
- Au, J.; Youngentob, K.N.; Foley, W.J.; Moore, B.D.; Fearn, T. Sample selection, calibration and validation of models developed from a large dataset of near infrared spectra of tree leaves. JNIRS 2020, 28, 186–203. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef] [Green Version]
- Fandango, A. Mastering TensorFlow 1. x: Advanced Machine Learning and Deep Learning Concepts Using TensorFlow 1. x and Keras; Packt Publishing Ltd.: Birmingham, UK, 2018. [Google Scholar]
- Savitzky, A.; Golay, M.J. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Hopkins, D.M. Using data pretreatments effectively. In Proceedings of the International Diffuse Reflectance Conference, Chambersburg, PA, USA, 3–8 August 2008. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Zhang, X.; Lin, T.; Xu, J.; Luo, X.; Ying, Y. DeepSpectra: An end-to-end deep learning approach for quantitative spectral analysis. Anal. Chim. Acta. 2019, 1058, 48–57. [Google Scholar] [CrossRef]
- Williams, P. The RPD statistic: A tutorial note. NIR News 2014, 25, 22–26. [Google Scholar] [CrossRef]
- Williams, P.; Dardenne, P.; Flinn, P. Tutorial: Items to be included in a report on a near infrared spectroscopy project. J. Near Infrared Spectrosc. 2017, 25, 85–90. [Google Scholar] [CrossRef]
- Williams, P.; Manley, M.; Antoniczyn, J. Near-InfraRed Technoloy-Getting the Best Out of Light; Sun Press Imprint: Stellenbosch, South Africa, 2019; p. 301. [Google Scholar]
- Kumar, A.; Tomer, V.; Kaur, A.; Kumar, V.; Gupta, K. Millets: A solution to agrarian and nutritional challenges. Agric. Food Secur. 2018, 7, 31. [Google Scholar] [CrossRef]
- Cozzolino, D. An overview of the use of infrared spectroscopy and chemometrics in authenticity and traceability of cereals. Food Res. Int. 2014, 60, 262–265. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Sha, S.; Rong, Z.; Chen, J.; He, Q.; Khan, D.M.; Zhu, S. Feasibility study of near infrared spectroscopy with variable selection for non-destructive determination of quality parameters in shell-intact cottonseed. Ind. Crops Prod. 2013, 43, 654–660. [Google Scholar] [CrossRef]






| Species | Number of Samples | Range of Protein (%, (g·100 g−1)) | Average of Protein (%, (g·100 g−1)) |
|---|---|---|---|
| Finger millet | 20 | 5.99–9.59 | 7.93 |
| Foxtail millet | 19 | 9.08–13.42 | 11.50 |
| Maize | 10 | 8.53–9.97 | 9.14 |
| Pearl millet | 125 | 9.69–21.51 | 15.78 |
| Sorghum | 154 | 8.68–18.38 | 13.09 |
| Multiple cereals | 328 | 5.99–21.51 | 13.59 |
| Instrument | Method | Set | Slope | Intercept | R2 | RMSE | RPD |
|---|---|---|---|---|---|---|---|
| FOSS-DS2500 | WinISI software | Calibration | 0.87 | 1.74 | 0.90 | 0.91 | 3.56 |
| Validation | 0.82 | 2.38 | 0.86 | 1.09 | 3.08 | ||
| Hone Create software | Calibration | 0.95 | 0.64 | 0.96 | 0.66 | 4.93 | |
| Validation | 0.89 | 1.44 | 0.90 | 1.00 | 3.38 | ||
| CNN-based customized pipeline | Calibration | 0.98 | 0.29 | 0.99 | 0.33 | 9.85 | |
| Validation | 0.88 | 1.61 | 0.89 | 1.03 | 3.26 | ||
| HL-EVT5 | Hone Create software | Calibration | 0.97 | 0.43 | 0.98 | 0.42 | 7.79 |
| Validation | 0.90 | 1.35 | 0.91 | 0.97 | 3.48 | ||
| CNN- based customized pipeline | Calibration | 0.98 | 0.28 | 0.98 | 0.46 | 7.00 | |
| Validation | 0.87 | 1.70 | 0.87 | 1.10 | 3.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chadalavada, K.; Anbazhagan, K.; Ndour, A.; Choudhary, S.; Palmer, W.; Flynn, J.R.; Mallayee, S.; Pothu, S.; Prasad, K.V.S.V.; Varijakshapanikar, P.; et al. NIR Instruments and Prediction Methods for Rapid Access to Grain Protein Content in Multiple Cereals. Sensors 2022, 22, 3710. https://doi.org/10.3390/s22103710
Chadalavada K, Anbazhagan K, Ndour A, Choudhary S, Palmer W, Flynn JR, Mallayee S, Pothu S, Prasad KVSV, Varijakshapanikar P, et al. NIR Instruments and Prediction Methods for Rapid Access to Grain Protein Content in Multiple Cereals. Sensors. 2022; 22(10):3710. https://doi.org/10.3390/s22103710
Chicago/Turabian StyleChadalavada, Keerthi, Krithika Anbazhagan, Adama Ndour, Sunita Choudhary, William Palmer, Jamie R. Flynn, Srikanth Mallayee, Sharada Pothu, Kodukula Venkata Subrahamanya Vara Prasad, Padmakumar Varijakshapanikar, and et al. 2022. "NIR Instruments and Prediction Methods for Rapid Access to Grain Protein Content in Multiple Cereals" Sensors 22, no. 10: 3710. https://doi.org/10.3390/s22103710