Hybrid Network with Attention Mechanism for Detection and Location of Myocardial Infarction Based on 12-Lead Electrocardiogram Signals
Abstract
:1. Introduction
- A novel multi-lead attention (MLA) mechanism integrated with CNN and bidirectional gated recurrent unit (BiGRU) framework (MLA-CNN-BiGRU) is proposed. The parallel deployed CNN and BiGRU modules are innovatively utilized to extract features to detect and locate MI via 12-lead heartbeat signals. As far as we know, this fills the gap of applying deep learning methods to automatically extract spatial and temporal features from 12-lead ECG signals in MI diagnosis. The proposed feature extraction method paves a new way for feature engineering.
- The MLA is developed by the designed activation function. The proposed attention mechanism measures and exploits the contribution of each lead to boost the diagnostic performance. Existing studies mainly focus on manual selection of leads or treat all the leads equally with repeated and redundant information. With the proposed model-based approach, this study serves as a preliminary exploration on the importance evaluation of each lead for MI detection and location.
- Different leads are interrelated and correlated. It is essential to fully exploit available features to enhance the performance. To our knowledge, it is the first time to adopt 2D-CNN to extract spatial features based on multi-lead fusion in MI diagnosis. Three different convolutional kernels are innovatively applied to extract correlation and regional features among different leads.
- MI detection and location under intra-patient and inter-patient schemes are all performed to test the robustness of MLA-CNN-BiGRU. In addition, elaborate and exhaustive ablation experiments are carried out to verify the effectiveness of the framework. Experimental results indicate that the proposed intelligent framework achieves satisfactory performance and demonstrates vital clinical significance.
2. Related Work
2.1. Attention Mechanism
2.2. Convolutional Neural Network
2.3. Gated Recurrent Unit
3. Dataset and Pre-Processing
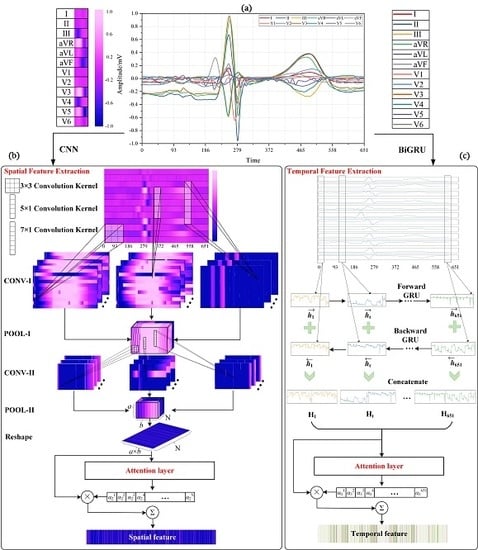
4. Methodology
4.1. Multi-lead Attention Module
4.2. CNN with Attention Mechanism for Spatial Feature Extraction
4.2.1. Convolutional Layer
4.2.2. Pooling Layer
4.2.3. Attention Layer for CNN
4.3. BiGRU with Attention Mechanism for Temporal Feature Extraction
4.3.1. BiGRU Neural Network
4.3.2. Attention Layer for BiGRU
4.4. Merge and Classification
| Algorithm 1 Training process of the proposed framework. |
| Input: PTB Dataset , Epoch E, Batch size B Output: The well-trained hybrid neural network
|
5. Results
5.1. Evaluation Metrics
5.2. Experimental Methodology
5.3. MI Detection
5.3.1. Intra-Patient Scheme
5.3.2. Inter-Patient Scheme
5.4. MI Location
5.4.1. Intra-Patient Scheme
5.4.2. Inter-Patient Scheme
6. Discussion
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Benjamin, E.J.; Muntner, P.; Bittencourt, M.S. Heart disease and stroke statistics—2019 update: A report from the American Heart Association. Circulation 2019, 139, e56–e528. [Google Scholar] [CrossRef] [PubMed]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Chaitman, B.R.; Bax, J.J.; Morrow, D.A.; White, H.D.; The Executive Group on behalf of the Joint European Society of Cardiology (ESC); American College of Cardiology (ACC); American Heart Association (AHA); et al. Fourth universal definition of myocardial infarction (2018). J. Am. Coll. Cardiol. 2018, 72, 2231–2264. [Google Scholar] [CrossRef] [PubMed]
- Sadhukhan, D.; Pal, S.; Mitra, M. Automated identification of myocardial infarction using harmonic phase distribution pattern of ECG data. IEEE Trans. Instrum. Meas. 2018, 67, 2303–2313. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J.; Wang, G.; Huang, K.; Li, F.; Zheng, Y.; Luo, Y.; Zhou, F. A novel electrocardiogram parameterization algorithm and its application in myocardial infarction detection. Comput. Biol. Med. 2015, 61, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Mixon, T.A.; Suhr, E.; Caldwell, G.; Greenberg, R.D.; Colato, F.; Blackwell, J.; Jo, C.H.; Dehmer, G.J. Retrospective description and analysis of consecutive catheterization laboratory ST-segment elevation myocardial infarction activations with proposal, rationale, and use of a new classification scheme. Circ. Cardiovasc. Qual. Outcomes 2012, 5, 62–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faust, O.; Acharya, U.R.; Tamura, T. Formal design methods for reliable computer-aided diagnosis: A review. IEEE Rev. Biomed. Eng. 2012, 5, 15–28. [Google Scholar] [CrossRef]
- Lu, H.; Ong, K.; Chia, P. An automated ECG classification system based on a neuro-fuzzy system. In Proceedings of the Computers in Cardiology 2000, Cambridge, MA, USA, 24–27 September 2000; pp. 387–390. [Google Scholar]
- Ansari, S.; Farzaneh, N.; Duda, M.; Horan, K.; Andersson, H.B.; Goldberger, Z.D.; Nallamothu, B.K.; Najarian, K. A review of automated methods for detection of myocardial ischemia and infarction using electrocardiogram and electronic health records. IEEE Rev. Biomed. Eng. 2017, 10, 264–298. [Google Scholar] [CrossRef]
- Barmpoutis, P.; Dimitropoulos, K.; Apostolidis, A.; Grammalidis, N. Multi-lead ECG signal analysis for myocardial infarction detection and localization through the mapping of Grassmannian and Euclidean features into a common Hilbert space. Biomed. Signal Process. Control 2019, 52, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Mitra, M. Application of cross wavelet transform for ECG pattern analysis and classification. IEEE Trans. Instrum. Meas. 2013, 63, 326–333. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Adam, M.; Lih, O.S.; Sudarshan, V.K.; Hong, T.J.; Koh, J.E.; Hagiwara, Y.; Chua, C.K.; Poo, C.K.; et al. Automated characterization and classification of coronary artery disease and myocardial infarction by decomposition of ECG signals: A comparative study. Inf. Sci. 2017, 377, 17–29. [Google Scholar] [CrossRef]
- Kumar, M.; Pachori, R.; Acharya, U. Automated diagnosis of myocardial infarction ECG signals using sample entropy in flexible analytic wavelet transform framework. Entropy 2017, 19, 488. [Google Scholar] [CrossRef]
- Sharma, L.; Tripathy, R.; Dandapat, S. Multiscale energy and eigenspace approach to detection and localization of myocardial infarction. IEEE Trans. Biomed. Eng. 2015, 62, 1827–1837. [Google Scholar] [CrossRef]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Fu, L.; Nie, B.; Peng, Z.; Liu, H. A Novel Framework with High Diagnostic Sensitivity for Lung Cancer Detection by Electronic Nose. Sensors 2019, 19, 5333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Jia, K. FusionAtt: Deep Fusional Attention Networks for Multi-Channel Biomedical Signals. Sensors 2019, 19, 2429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, D.; Wang, H.; Huang, X.; Gidlund, M. CarNet: A Dual Correlation Method for Health Perception of Rotating Machinery. IEEE Sens. J. 2019. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M. Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inf. Sci. 2017, 415, 190–198. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, M.; Zhang, Y.; Liao, Y.; Huang, Q.; Chang, S.; Wang, H.; He, J. Real-time multilead convolutional neural network for myocardial infarction detection. IEEE J. Biomed. Health Informat. 2017, 22, 1434–1444. [Google Scholar] [CrossRef]
- Lui, H.W.; Chow, K.L. Multiclass classification of myocardial infarction with convolutional and recurrent neural networks for portable ECG devices. Informat. Med. Unlocked 2018, 13, 26–33. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J. Application of Heartbeat-Attention Mechanism for Detection of Myocardial Infarction Using 12-Lead ECG Records. Appl. Sci. 2019, 9, 3328. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wang, F.; Huang, Q.; Chang, S.; Wang, H.; He, J. MFB-CBRNN: A hybrid network for MI detection using 12-lead ECGs. IEEE J. Biomed. Health Inform. 2019, 24, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Shi, L. ML–ResNet: A novel network to detect and locate myocardial infarction using 12 leads ECG. Comput. Methods Programs Biomed. 2020, 185, 105138. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Shi, L. Automated interpretable detection of myocardial infarction fusing energy entropy and morphological features. Comput. Methods Programs Biomed. 2019, 175, 9–23. [Google Scholar] [CrossRef]
- Itti, L.; Koch, C. Computational modelling of visual attention. Nat. Rev. Neurosci. 2001, 2, 194. [Google Scholar] [CrossRef] [Green Version]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems 30, Long Beach, CA, USA, 4–9 December 2017; pp. 5998–6008. [Google Scholar]
- Lin, Z.; Feng, M.; Santos, C.N.; Yu, M.; Xiang, B.; Zhou, B.; Bengio, Y. A structured self-attentive sentence embedding. In Proceedings of the 5th International Conference on Learning Representations, Toulon, France, 24–26 April 2017; pp. 448–456. [Google Scholar]
- Huang, T.; Deng, Z.H.; Shen, G.; Chen, X. A Window-Based Self-Attention approach for sentence encoding. Neurocomputing 2020, 375, 25–31. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: an overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef] [Green Version]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32th International Conference on Machine Learning, Lille, France, 6 July–11 July 2015; pp. 448–456. [Google Scholar]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving neural networks by preventing co-adaptation of feature detectors. arXiv 2012, arXiv:1207.0580. [Google Scholar]
- Liu, N.; Wang, L.; Chang, Q.; Xing, Y.; Zhou, X. A Simple and Effective Method for Detecting Myocardial Infarction Based on Deep Convolutional Neural Network. J. Med. Imaging Health Informat. 2018, 8, 1508–1512. [Google Scholar] [CrossRef]
- Baloglu, U.B.; Talo, M.; Yildirim, O.; San Tan, R.; Acharya, U.R. Classification of myocardial infarction with multi-lead ECG signals and deep CNN. Pattern Recognit. Lett. 2019, 122, 23–30. [Google Scholar] [CrossRef]
- Liu, W.; Huang, Q.; Chang, S.; Wang, H.; He, J. Multiple-feature-branch convolutional neural network for myocardial infarction diagnosis using electrocardiogram. Biomed. Signal Process. Control 2018, 45, 22–32. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Van Merriënboer, B.; Bahdanau, D.; Bengio, Y. On the properties of neural machine translation: Encoder-decoder approaches. arXiv 2014, arXiv:1409.1259. [Google Scholar]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural machine translation by jointly learning to align and translate. arXiv 2014, arXiv:1409.0473. [Google Scholar]
- Chen, J.; Jiang, D.; Zhang, Y. A Hierarchical Bidirectional GRU Model With Attention for EEG-Based Emotion Classification. IEEE Access 2019, 7, 118530–118540. [Google Scholar] [CrossRef]
- Lynn, H.M.; Pan, S.B.; Kim, P. A Deep Bidirectional GRU Network Model for Biometric Electrocardiogram Classification Based on Recurrent Neural Networks. IEEE Access 2019, 7, 145395–145405. [Google Scholar] [CrossRef]
- PhysioBank, P. PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar]
- Martis, R.J.; Acharya, U.R.; Min, L.C. ECG beat classification using PCA, LDA, ICA and discrete wavelet transform. Biomed. Signal Process. Control 2013, 8, 437–448. [Google Scholar] [CrossRef]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Wang, T.; Zhao, M.; Wang, P. An adaptive multi-sensor data fusion method based on deep convolutional neural networks for fault diagnosis of planetary gearbox. Sensors 2017, 17, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boureau, Y.L.; Ponce, J.; LeCun, Y. A theoretical analysis of feature pooling in visual recognition. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010; pp. 111–118. [Google Scholar]
- Zhang, G.; Tang, L.; Zhou, L.; Liu, Z.; Liu, Y.; Jiang, Z. Principal Component Analysis Method with Space and Time Windows for Damage Detection. Sensors 2019, 19, 2521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, P.C.; Lin, J.J.; Hsieh, J.C.; Weng, J. Myocardial infarction classification with multi-lead ECG using hidden Markov models and Gaussian mixture models. Appl. Soft Comput. 2012, 12, 3165–3175. [Google Scholar] [CrossRef]
- Crawford, M.H.; Bernstein, S.J.; Deedwania, P.C.; DiMarco, J.P.; Ferrick, K.J.; Garson, A.; Green, L.A.; Greene, H.L.; Silka, M.J.; Stone, P.H.; et al. ACC/AHA Guidelines for Ambulatory Electrocardiography: Executive Summary and Recommendations. Circulation 1999, 100, 886–893. [Google Scholar] [CrossRef] [Green Version]
- Acharya, U.R.; Fujita, H.; Sudarshan, V.K.; Oh, S.L.; Adam, M.; Koh, J.E.; Tan, J.H.; Ghista, D.N.; Martis, R.J.; Chua, C.K.; et al. Automated detection and localization of myocardial infarction using electrocardiogram: A comparative study of different leads. Knowl.-Based Syst. 2016, 99, 146–156. [Google Scholar] [CrossRef]





| Class | No. of Records | No. of 12-Lead Beats |
|---|---|---|
| AMI | 47 | 81,168 |
| ALMI | 43 | 80,988 |
| ASMI | 79 | 140,256 |
| IMI | 89 | 151,716 |
| ILMI | 56 | 97,296 |
| Other MIs | 54 | 81,516 |
| HCs | 80 | 127,188 |
| Total | 448 | 760,128 |
| MLA-BiGRU | Acc (%) | Sen (%) | Spe (%) | MLA-CNN | Acc (%) | Sen (%) | Spe (%) |
|---|---|---|---|---|---|---|---|
| Fold 1 | 86.90 | 96.06 | 41.32 | Fold 1 | 91.81 | 97.28 | 64.62 |
| Fold 2 | 91.58 | 96.19 | 68.56 | Fold 2 | 92.16 | 98.80 | 59.05 |
| Fold 3 | 93.46 | 97.97 | 70.75 | Fold 3 | 93.41 | 96.31 | 78.77 |
| Fold 4 | 87.84 | 96.62 | 45.30 | Fold 4 | 92.94 | 99.29 | 62.18 |
| Fold 5 | 96.24 | 97.37 | 90.71 | Fold 5 | 87.51 | 99.34 | 29.37 |
| Mean | 91.20 | 96.84 | 63.33 | Mean | 91.57 | 98.20 | 58.80 |
| Std | 3.89 | 0.81 | 20.26 | Std | 2.35 | 1.35 | 18.10 |
| MLA-BiGRU | Acc (%) | Sen (%) | Spe (%) | MLA-CNN | Acc (%) | Sen (%) | Spe (%) |
| Fold 1 | 96.43 | 98.29 | 87.17 | Fold 1 | 93.91 | 100.00 | 63.63 |
| Fold 2 | 83.31 | 100.00 | 0.00 | Fold 2 | 91.69 | 99.75 | 51.44 |
| Fold 3 | 95.50 | 99.13 | 77.19 | Fold 3 | 99.61 | 99.80 | 98.66 |
| Fold 4 | 99.62 | 99.61 | 99.68 | Fold 4 | 99.73 | 99.99 | 98.48 |
| Fold 5 | 91.94 | 91.91 | 92.11 | Fold 5 | 99.84 | 99.88 | 99.67 |
| Mean | 93.36 | 97.79 | 71.23 | Mean | 96.96 | 99.88 | 82.38 |
| Std | 6.25 | 3.35 | 40.65 | Std | 3.88 | 0.11 | 23.09 |
| CNN-BiGRU | Acc (%) | Sen (%) | Spe (%) | MLA-CNN-BiGRU | Acc (%) | Sen (%) | Spe (%) |
| Fold 1 | 97.64 | 99.31 | 89.34 | Fold 1 | 99.93 | 99.99 | 99.62 |
| Fold 2 | 98.27 | 99.46 | 92.34 | Fold 2 | 99.85 | 100.00 | 99.10 |
| Fold 3 | 98.31 | 99.70 | 91.32 | Fold 3 | 99.95 | 99.99 | 99.76 |
| Fold 4 | 91.06 | 99.26 | 51.34 | Fold 4 | 99.96 | 99.99 | 99.82 |
| Fold 5 | 93.54 | 98.92 | 67.09 | Fold 5 | 99.97 | 99.99 | 99.86 |
| Mean | 95.76 | 99.33 | 78.29 | Mean | 99.93 | 99.99 | 99.63 |
| Std | 3.29 | 0.29 | 18.31 | Std | 0.05 | 0.004 | 0.31 |
| Framework | Intra-Patient Scheme | Inter-Patient Scheme | ||||||
|---|---|---|---|---|---|---|---|---|
| Folds | Acc (%) | Sen (%) | Spe (%) | Folds | Acc (%) | Sen (%) | Spe (%) | |
| PCA-MLP | Fold 1 | 72.45 | 85.70 | 6.56 | Fold 1 | 79.38 | 91.43 | 25.16 |
| Fold 2 | 76.61 | 89.85 | 10.54 | Fold 2 | 54.11 | 76.32 | 12.13 | |
| Fold 3 | 74.69 | 86.90 | 13.07 | Fold 3 | 68.72 | 81.25 | 0.76 | |
| Fold 4 | 89.72 | 97.16 | 53.69 | Fold 4 | 78.70 | 84.79 | 0.00 | |
| Fold 5 | 91.48 | 96.96 | 64.52 | Fold 5 | 77.20 | 91.47 | 0.00 | |
| Mean | 80.99 | 91.31 | 29.68 | Mean | 71.62 | 85.05 | 7.61 | |
| Std | 8.92 | 5.46 | 27.24 | Std | 10.68 | 6.57 | 11.08 | |
| MLA-CNN-BiGRU | Mean | 99.93 | 99.99 | 99.63 | Mean | 96.50 | 97.10 | 93.34 |
| Std | 0.05 | 0.004 | 0.31 | Std | 2.25 | 2.60 | 4.84 | |
| MLA-BiGRU | Acc (%) | Sen (%) | Spe (%) | MLA-CNN | Acc (%) | Sen (%) | Spe (%) |
|---|---|---|---|---|---|---|---|
| Fold 1 | 80.98 | 92.15 | 30.71 | Fold 1 | 87.04 | 94.23 | 54.73 |
| Fold 2 | 87.11 | 83.95 | 93.10 | Fold 2 | 85.99 | 86.68 | 84.70 |
| Fold 3 | 85.56 | 92.62 | 47.25 | Fold 3 | 85.74 | 99.88 | 9.02 |
| Fold 4 | 92.52 | 95.86 | 49.41 | Fold 4 | 91.31 | 92.09 | 81.24 |
| Fold 5 | 84.40 | 100.00 | 0.00 | Fold 5 | 90.70 | 97.21 | 55.47 |
| Mean | 86.11 | 92.92 | 44.09 | Mean | 88.16 | 94.02 | 57.03 |
| Std | 4.23 | 5.91 | 33.78 | Std | 2.65 | 5.05 | 30.27 |
| MLA-BiGRU | Acc (%) | Sen (%) | Spe (%) | MLA-CNN | Acc (%) | Sen (%) | Spe (%) |
| Fold 1 | 84.83 | 94.54 | 41.11 | Fold 1 | 90.47 | 99.97 | 47.72 |
| Fold 2 | 89.59 | 84.24 | 99.69 | Fold 2 | 93.83 | 94.34 | 92.85 |
| Fold 3 | 84.44 | 100.00 | 0.00 | Fold 3 | 95.59 | 100.00 | 71.65 |
| Fold 4 | 93.20 | 99.97 | 5.70 | Fold 4 | 93.07 | 99.99 | 3.68 |
| Fold 5 | 86.19 | 99.99 | 11.52 | Fold 5 | 99.90 | 100.00 | 99.36 |
| Mean | 87.65 | 95.75 | 31.60 | Mean | 94.57 | 98.86 | 63.05 |
| Std | 3.71 | 6.85 | 41.23 | Std | 3.50 | 2.53 | 38.86 |
| CNN-BiGRU | Acc (%) | Sen (%) | Spe (%) | MLA-CNN-BiGRU | Acc (%) | Sen (%) | Spe (%) |
| Fold 1 | 93.69 | 95.71 | 84.58 | Fold 1 | 92.93 | 93.70 | 89.48 |
| Fold 2 | 97.29 | 98.59 | 94.84 | Fold 2 | 95.59 | 95.20 | 96.33 |
| Fold 3 | 88.97 | 99.97 | 29.25 | Fold 3 | 97.93 | 98.92 | 92.55 |
| Fold 4 | 96.18 | 96.61 | 90.62 | Fold 4 | 97.87 | 97.70 | 100.00 |
| Fold 5 | 86.07 | 99.97 | 10.89 | Fold 5 | 98.17 | 99.98 | 88.36 |
| Mean | 92.44 | 98.17 | 62.04 | Mean | 96.50 | 97.10 | 93.34 |
| Std | 4.79 | 1.95 | 39.03 | Std | 2.25 | 2.60 | 4.84 |
| Folds | Category | Intra-patient Scheme | Inter-patient Scheme | ||||
|---|---|---|---|---|---|---|---|
| Acc (%) | Sen (%) | Spe (%) | Acc (%) | Sen (%) | Spe (%) | ||
| Fold 1 | AMI | 98.13 | 99.70 | 97.93 | 62.06 | 78.51 | 59.31 |
| ALMI | 98.13 | 96.97 | 98.30 | 62.06 | 22.78 | 66.05 | |
| ASMI | 98.13 | 93.64 | 99.29 | 62.06 | 58.90 | 63.02 | |
| IMI | 98.13 | 99.80 | 97.65 | 62.06 | 41.64 | 66.28 | |
| ILMI | 98.13 | 99.38 | 97.93 | 62.06 | 58.18 | 62.93 | |
| HC | 98.13 | 99.86 | 97.74 | 62.06 | 97.24 | 54.51 | |
| Mean | 98.13 | 98.22 | 98.14 | 62.06 | 59.54 | 62.02 | |
| Fold 2 | AMI | 98.07 | 93.74 | 98.64 | 58.61 | 39.87 | 61.20 |
| ALMI | 98.07 | 95.81 | 98.38 | 58.61 | 54.53 | 58.86 | |
| ASMI | 98.07 | 97.05 | 98.34 | 58.61 | 35.45 | 65.90 | |
| IMI | 98.07 | 99.76 | 97.58 | 58.61 | 82.28 | 52.59 | |
| ILMI | 98.07 | 99.82 | 97.78 | 58.61 | 67.09 | 56.43 | |
| HC | 98.07 | 99.95 | 97.64 | 58.61 | 67.19 | 56.79 | |
| Mean | 98.07 | 97.69 | 98.06 | 58.61 | 57.74 | 58.63 | |
| Fold 3 | AMI | 99.73 | 99.78 | 99.72 | 46.19 | 89.88 | 39.87 |
| ALMI | 99.73 | 98.59 | 99.88 | 46.19 | 99.68 | 44.66 | |
| ASMI | 99.73 | 99.96 | 99.67 | 46.19 | 12.77 | 65.39 | |
| IMI | 99.73 | 99.88 | 99.68 | 46.19 | 72.31 | 42.72 | |
| ILMI | 99.73 | 99.75 | 99.72 | 46.19 | 34.19 | 48.60 | |
| HC | 99.73 | 99.95 | 99.67 | 46.19 | 67.29 | 41.04 | |
| Mean | 99.73 | 99.65 | 99.72 | 46.19 | 62.69 | 47.05 | |
| Fold 4 | AMI | 99.85 | 99.78 | 99.86 | 72.68 | 72.64 | 72.69 |
| ALMI | 99.85 | 99.79 | 99.86 | 72.68 | 65.10 | 74.03 | |
| ASMI | 99.85 | 99.96 | 99.82 | 72.68 | 46.56 | 75.98 | |
| IMI | 99.85 | 99.80 | 99.86 | 72.68 | 81.03 | 69.84 | |
| ILMI | 99.85 | 99.75 | 99.87 | 72.68 | 95.48 | 70.34 | |
| HC | 99.85 | 99.95 | 99.82 | 72.68 | 71.59 | 72.85 | |
| Mean | 99.85 | 99.84 | 99.85 | 72.68 | 72.07 | 72.62 | |
| Fold 5 | AMI | 99.75 | 99.78 | 99.75 | 75.18 | 57.96 | 78.53 |
| ALMI | 99.75 | 99.54 | 99.78 | 75.18 | 100.00 | 74.13 | |
| ASMI | 99.75 | 100.00 | 99.69 | 75.18 | 69.05 | 75.91 | |
| IMI | 99.75 | 99.96 | 99.69 | 75.18 | 93.48 | 66.01 | |
| ILMI | 99.75 | 99.15 | 99.86 | 75.18 | 2.28 | 81.78 | |
| HC | 99.75 | 99.81 | 99.74 | 75.18 | 83.99 | 71.86 | |
| Mean | 99.75 | 99.71 | 99.75 | 75.18 | 67.79 | 74.70 | |
| five-fold Mean | \ | 99.11 | 99.02 | 99.10 | 62.94 | 63.97 | 63.00 |
| Year | Lead* | Records or Beats | Dataset | Framework | Detection | Location | Performance | |
|---|---|---|---|---|---|---|---|---|
| Intra-Patient | Inter-Patient | |||||||
| 2016 [51] | Lead 11 for detection (V5) Lead 9 for location (V3) | Beats | 485,753 MI 125,652 HC | DWT + KNN | ✓ | ✓ | Detection: Acc = 98.80% Sen = 99.45% Spe = 96.27% Location: Acc = 98.74% Sen = 99.55% Spe = 99.16% | No |
| 2017 [12] | Lead 2 (II) | Beats | 40,182 MI 10,546 HC | FAWT and SEnt + LS-SVM | ✓ | × | Acc = 99.31% Sen = 99.62% Spe = 98.12% | No |
| 2017 [19] | Lead 2 (II) | Beats | 40,182 MI 10,546 HC | CNN | ✓ | × | Acc = 95.22% Sen = 95.49% Spe = 94.19% | No |
| 2017 [20] | Lead 5, 8, 9 and 11 (aVL, V2, V3 and V5) | Beats | 167 MI records 80 HC records | ML-CNN | ✓ | × | Acc = 96.00% Sen = 95.40% Spe = 97.37% | No |
| 2018 [3] | Lead 2,3 and 8 (II, III, and V2) | Beats | 15,000 MI 5000 HC | Handcrafted features + LR | ✓ | × | Acc = 95.60% Sen = 96.50% Spe = 92.70% | No |
| 2018 [21] | Lead 1 (I) | Records | 368 MI 80 HC 74 Other 278 Noisy | CNN-LSTM stacking decoding | ✓ | × | No | Sen = 92.4% Spe = 97.7% |
| 2019 [22] | 12 Leads | Records | 369 MI 79 HC | BiLSTM Heartbeat-attention | ✓ | × | No | Acc = 94.77% Sen = 95.58% Spe = 90.48% |
| 2019 [25] | 12 Leads | Beats | 28,213 MI 5373 HC | MODWPT + PCA + SVM (Intra) MODWPT + PCA + Bagging (Inter) | ✓ | × | Acc = 99.75% Sen = 99.37% Spe = 99.37% | Acc = 92.69% Sen = 80.96% Spe = 80.96% |
| 2019 [23] | 12 Leads | Beats | 53,712 MI 10,638 HC | CNN + BiLSTM | ✓ | × | Acc = 99.90% Sen = 99.97% Spe = 99.54% | Acc = 93.08% Sen = 94.42% Spe = 86.29% |
| 2019 [24] | 12 Leads | Beats | 28,213MI 5373 HC | ML-ResNet | ✓ | ✓ | Detection: Acc = 99.92% Sen = 99.98% Spe = 99.77% Location: Acc = 99.72% Sen = 99.63% Spe = 99.72% | Detection: Acc = 95.49% Sen = 94.85% Spe = 97.37% Location: Acc = 55.74% Sen = 47.58% Spe = 55.37% |
| Proposed | 12 Leads | Beats | 632,940 MI 127,188 HC | MLA-CNN-BiGRU | ✓ | ✓ | Detection: Acc = 99.93% Sen = 99.99% Spe = 99.63% Location: Acc = 99.11% Sen = 99.02% Spe = 99.10% | Detection: Acc = 96.50% Sen = 97.10% Spe = 93.34% Location: Acc = 62.94% Sen = 63.97% Spe = 63.00% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, L.; Lu, B.; Nie, B.; Peng, Z.; Liu, H.; Pi, X. Hybrid Network with Attention Mechanism for Detection and Location of Myocardial Infarction Based on 12-Lead Electrocardiogram Signals. Sensors 2020, 20, 1020. https://doi.org/10.3390/s20041020
Fu L, Lu B, Nie B, Peng Z, Liu H, Pi X. Hybrid Network with Attention Mechanism for Detection and Location of Myocardial Infarction Based on 12-Lead Electrocardiogram Signals. Sensors. 2020; 20(4):1020. https://doi.org/10.3390/s20041020
Chicago/Turabian StyleFu, Lidan, Binchun Lu, Bo Nie, Zhiyun Peng, Hongying Liu, and Xitian Pi. 2020. "Hybrid Network with Attention Mechanism for Detection and Location of Myocardial Infarction Based on 12-Lead Electrocardiogram Signals" Sensors 20, no. 4: 1020. https://doi.org/10.3390/s20041020