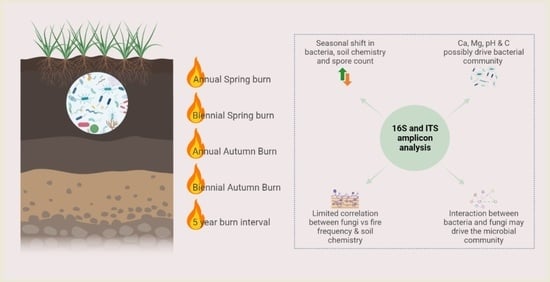
Microbial Community Responses to Alterations in Historical Fire Regimes in Montane Grasslands
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Sampling and Soil Chemical Analysis
2.3. Mycorrhizal Assessment
2.4. DNA Extraction, Amplification, and Sequencing
2.5. Bioinformatic Analysis
2.6. Statistical Analysis
3. Results
3.1. Soil Chemistry
3.2. Microbial Community Composition of Burn-Treated Soils
3.2.1. Soil Bacterial Community Profile
3.2.2. Fungal Community Profile of the Soil
3.3. Effect of Soil Chemistry on Microbial Community Composition
4. Discussion
4.1. Correlation of Bacterial Community Composition with Fire Event Frequency
4.2. Fungal Communities and Fire Event Frequency
4.3. Impact of Soil Chemistry on Microbial Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bond, W.J. Open Ecosystems: Ecology and Evolution beyond the Forest Edges, online ed.; Oxford University Press: Oxford, UK, 2019. [Google Scholar]
- Bond, W.J.; Keeley, J.E. Fire as a global ‘herbivore’: The ecology and evolution of flammable ecosystems. Trends Ecol. Evol. 2005, 20, 387–394. [Google Scholar] [CrossRef]
- Coop, J.D.; Givnish, T.J. Spatial and temporal patterns of recent forest encroachment in montane grasslands of the Valles Caldera, New Mexico, USA. J. Biogeogr. 2007, 34, 914–927. [Google Scholar] [CrossRef]
- Carilla, J.; Grau, H.R. 150 years of tree establishment, land use and climate change in montane grasslands, northwest Argentina. Biotropica 2010, 42, 49–58. [Google Scholar] [CrossRef]
- Oliver, V.; Oliveras, I.; Kala, J.; Lever, R.; Teh, Y.A. The effects of burning and grazing on soil carbon dynamics in managed Peruvian tropical montane grasslands. Biogeosciences 2017, 14, 5633–5646. [Google Scholar] [CrossRef] [Green Version]
- Suazo, M.M.; Collins, S.L.; Parmenter, R.R.; Muldavin, E. Montane valley grasslands are highly resistant to summer wildfire. J. Veg. Sci. 2018, 29, 1017–1028. [Google Scholar] [CrossRef]
- Bengtsson, J.; Bullock, J.M.; Egoh, B.; Everson, C.; Everson, T.; O’Connor, T.; O’Farrell, P.J.; Smith, H.G.; Lindborg, Y. Grasslands—More important for ecosystem services than you might think. Ecosphere 2019, 10, e02582. [Google Scholar] [CrossRef]
- Morris, C.D.; Everson, C.S.; Everson, T.M.; Gordijn, P.J. Frequent burning maintained a stable grassland over four decades in the Drakensberg, South Africa. Afr. J. Range Forage Sci. 2020, 38, 39–52. [Google Scholar] [CrossRef]
- Archibald, S.; Bond, W.J.; Stock, W.D.; Fairbanks, D.H.K. Shaping the landscape: Fire–grazer interactions in an African savanna. Ecol. Appl. 2005, 15, 96–109. [Google Scholar] [CrossRef]
- Beerling, D.J.; Osborne, C.P. The origin of the savanna biome. Glob. Chang. Biol. 2006, 12, 2023–2031. [Google Scholar] [CrossRef]
- Sinclair, A.R.; Metzger, K.L.; Mduma, S.; Fryxell, J.M. Serengeti IV: Sustaining Biodiversity in a Coupled Human-Natural System; University of Chicago Press: Chicago, IL, USA, 2012. [Google Scholar]
- Holdo, R.M.; Holt, R.D. African savannas. In Reference Module in Earth Systems and Environmental Sciences; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Klink, C.A.; Machado, R.B. Conservation of the Brazilian Cerrado. Conserv. Biol. 2005, 19, 707–713. [Google Scholar] [CrossRef]
- Oliveira, P.S.; Marquis, R.J. The Cerrados of Brazil: Ecology and Natural History of a Neotropical Savanna; Columbia University Press: New York, NY, USA, 2002. [Google Scholar]
- Paruelo, J.M.; Lauenroth, W.K. Perspectives on ecological research in the Argentine Pampas. Austral Ecol. 2008, 33, 343–362. [Google Scholar]
- O’Connor, T.G.; Uys, R.G.; Mills, A.J. Ecological effects of fire-breaks in the montane grasslands of the southern Drakensberg, South Africa. Afr. J. Range Forage Sci. 2004, 21, 1–9. [Google Scholar] [CrossRef]
- Manson, A.D.; Jewitt, D.; Short, A.D. Effect of season and frequency of burning on soils and landscape functioning in a moist montane grassland. Afr. J. Range Forage Sci. 2007, 24, 9–18. [Google Scholar] [CrossRef]
- Everson, C.S.; Everson, T.M. The long-term effects of fire regime on primary production of montane grasslands in South Africa. Afr. J. Range Forage Sci. 2016, 33, 33–41. [Google Scholar] [CrossRef]
- McGranahan, D.A.; Archibald, S.; Kirkman, K.P.; O’Connor, T.G. A native C3 grass alters fuels and fire spread in montane grassland of South Africa. Plant Ecol. 2018, 219, 621–632. [Google Scholar] [CrossRef]
- Parr, C.L.; Lehmann, C.E.R.; Bond, W.J.; Hoffmann, W.A.; Andersen, A.N. Tropical grassy biomes: Misunderstood, neglected, and under threat. Trends Ecol. Evol. 2014, 29, 205–213. [Google Scholar] [CrossRef]
- Pressler, Y.; Moore, J.C.; Cotrufo, M.F. Belowground community responses to fire: Meta-analysis reveals contrasting responses of soil microorganisms and mesofauna. Oikos 2019, 128, 309–327. [Google Scholar] [CrossRef]
- Qin, C.; Zhu, K.; Chiariello, N.R.; Field, C.B.; Peay, K.G. Fire history and plant community composition outweigh decadal multi-factor global change as drivers of microbial composition in an annual grassland. J. Ecol. 2020, 108, 611–625. [Google Scholar] [CrossRef]
- Hooper, D.U.; Bignell, D.E.; Brown, V.K.; Brussard, L.; Dangerfield, J.M.; Wall, D.H.; Wardle, D.A.; Coleman, D.C.; Giller, K.E.; Lavelle, P.; et al. Interactions between Aboveground and Belowground Biodiversity in Terrestrial Ecosystems: Patterns, Mechanisms, and Feedbacks: We assess the evidence for correlation between aboveground and belowground diversity and conclude that a variety of mechanisms could lead to positive, negative, or no relationship-depending on the strength and type of interactions among species. BioScience 2000, 50, 1049–1061. [Google Scholar]
- Vermeire, M.-L.; Thoresen, J.; Lennard, K.; Vikram, S.; Kirkman, K.; Swemmer, A.M.; Te Beest, M.; Siebert, F.; Gordijn, P.; Venter, Z.; et al. Fire and herbivory drive fungal and bacterial communities through distinct above- and belowground mechanisms. Sci. Total Environ. 2021, 785, 147189. [Google Scholar] [CrossRef]
- He, T.; Lamont, B.B.; Pausas, J.G. Fire as a key driver of Earth’s biodiversity. Biol. Rev. 2019, 94, 1983–2010. [Google Scholar] [CrossRef] [PubMed]
- Baldrian, P. Forest microbiome: Diversity, complexity and dynamics. FEMS Microbiol. Rev. 2017, 41, 109–130. [Google Scholar] [CrossRef] [Green Version]
- Wright, B.R.; Clarke, P.J. Relationships between soil temperatures and properties of fire in feathertop spinifex (Triodia schinzii (Henrard) Lazarides) sandridge desert in central Australia. Rangel. J. 2008, 30, 317–325. [Google Scholar] [CrossRef]
- Beringer, J.; Hutley, L.B.; Tapper, N.J.; Coutts, A.; Kerley, A.; O’Grady, A.P. Fire impacts on surface heat, moisture and carbon fluxes from a tropical savanna in northern Australia. Int. J. Wildland Fire 2003, 12, 333–340. [Google Scholar] [CrossRef]
- Mucina, L.; Rutherford, M.C. The Vegetation of South Africa, Lesotho and Swaziland. In Strelitzia 19; South African National Biodiversity Institute: Pretoria, South Africa, 2006. [Google Scholar]
- Acocks, J.P.H. Veld Types of South Africa, 3rd. ed. In Memoirs of the Botanical Survey of South Africa; South African National Biodiversity Institute: Pretoria, South Africa, 1988; Volume 57, pp. 1–146. [Google Scholar]
- Jourdan, F.; Feraud, G.; Bertrand, H.; Kampunzu, A.B.; Tshoso, G.; Watkeys, M.K.; Le Gall, B. Karoo large igneous province: Brevity, origin, and relation to mass extinction questioned by new Ar-40/Ar-39 age data. Geology 2005, 33, 745–748. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, B.A. Methods for the Determination of Total Organic Carbon in Soils and Sediments; United States Environmental Protection Agency: Washington, DC, USA, 2002. [Google Scholar]
- Smith, S.; Dickson, S. VA Mycorrhizas: Basic Research Techniques; Cooperative Research Centre for Soil and Land Management: Adelaide, Australia, 1997.
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Matcher, G.F.; Dorrington, R.A.; Henninger, T.O.; Froneman, P.W. Insights into the bacterial diversity in a freshwater-deprived permanently open Eastern Cape estuary, using 16S rRNA pyrosequencing analysis. Water SA 2011, 37, 381–390. [Google Scholar] [CrossRef] [Green Version]
- van Geel, M.; Busschaert, P.; Honnay, O.; Lievens, B. Evaluation of six primer pairs targeting the nuclear rRNA operon for characterization of arbuscular mycorrhizal fungal (AMF) communities using 454 pyrosequencing. J. Microbiol. Methods 2014, 106, 93–100. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Öpik, M.; Vanatoa, A.; Vanatoa, E.; Moora, M.; Davison, J.; Kalwij, J.M.; Reier, Ü.; Zobel, M. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol. 2010, 188, 223–241. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 4, D733–D745. [Google Scholar] [CrossRef] [Green Version]
- Clarke, K.R.; Gorley, R.N. PRIMER v7: User Manual/Tutorial; PRIMER-E: Plymouth, UK, 2015; p. 296. [Google Scholar]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moyes, A.B.; Kueppers, L.M.; Pett-Ridge, J.; Carper, D.L.; Vandehey, N.; O’Neil, J.; Frank, A.C. Evidence for foliar endophytic nitrogen fixation in a widely distributed subalpine conifer. New Phytol. 2016, 210, 657–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santalahti, M.; Sun, H.; Jumpponen, A.; Pennanen, T.; Heinonsalo, J. Vertical and seasonal dynamics of fungal communities in boreal scots pine forest soil. FEMS Microbiol. Ecol. 2016, 92, fiw141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, B.S.; Hallett, P.D.; Kuan, H.L.; Gregory, A.S.; Watts, C.W.; Whitmore, A.P. Functional resilience of soil microbial communities depends on both soil structure and microbial community composition. Biol. Fertil. Soils 2008, 44, 745–754. [Google Scholar] [CrossRef]
- Weber, C.F.; Lockhart, J.S.; Charaska, E.; Aho, K.; Lohse, K.A. Bacterial composition of soils in ponderosa pine and mixed conifer forests exposed to different wildfire burn severity. Soil Biol. Biochem. 2014, 69, 242–250. [Google Scholar] [CrossRef]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Hervé, V.; Labbé, J.; Lastovetsky, O.A.; et al. Bacterial-fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dangi, S.R.; Stahl, P.D.; Pendall, E.; Cleary, E.M.; Buyer, J.S. Recovery of soil microbial community structure after fire in a sagebrush-grassland ecosystem. Land Degrad. Dev. 2010, 21, 423–432. [Google Scholar] [CrossRef]
- Docherty, K.M.; Balser, T.C.; Bohannan, B.J.M.; Gutknecht, L.M. Soil microbial responses to fire and interacting global change factors in a California annual grassland. Biogeochemistry 2012, 109, 63–83. [Google Scholar] [CrossRef]
- Dooley, S.R.; Treseder, K.K. The effect of fire on microbial biomass: A meta-analysis of field studies. Biogeochemistry 2012, 109, 49–61. [Google Scholar] [CrossRef] [Green Version]
- Goberna, M.; Garcia, C.; Insam, H.; Hernández, M.T.; Verdú, M. Burning fire-prone Mediterranean shrublands: Immediate changes in soil microbial community structure and ecosystem functions. Microb. Ecol. 2012, 64, 242–255. [Google Scholar] [CrossRef]
- Ferrenberg, S.; O’Neill, S.P.; Knelman, J.E.; Todd, B.; Duggan, S.; Bradley, D.; Robinson, T.; Schmidt, S.K.; Townsend, A.R.; Williams, M.W.; et al. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J. 2013, 7, 1102–1111. [Google Scholar] [CrossRef] [Green Version]
- Patel, J.; Grab, S.; De Maayer, P. Distinct microbial communities across a climatically versatile summit in the Lesotho highlands. Ecol. Evol. 2023, 13, e9891. [Google Scholar] [CrossRef]
- Siles, J.A.; Margesin, R. Abundance and diversity of bacterial, archaeal, and fungal communities along an altitudinal gradient in alpine Forest soils: What are the driving factors? Microb. Ecol. 2016, 72, 207–220. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Si, G.; Wang, J.; Luo, T.; Zhang, G. Bacterial community in alpine grasslands along an altitudinal gradient on the Tibetan Plateau. FEMS Microbiol. Ecol. 2014, 87, 121–132. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Cong, J.; Lu, H.; Li, G.; Xue, Y.; Deng, Y.; Li, H.; Zhou, J.; Li, D. Soil bacterial diversity patterns and drivers along an elevational gradient on Shennongjia Mountain, China. Microb. Biotechnol. 2015, 8, 739–746. [Google Scholar] [CrossRef] [Green Version]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef] [Green Version]
- Kindler, R.; Miltner, A.; Thullner, M.; Richnow, H.-H.; Kästner, M. Fate of bacterial biomass derived fatty acids in soil and their contribution to soil organic matter. Org. Geochem. 2009, 40, 29–37. [Google Scholar] [CrossRef]
- Khodadad, C.L.M.; Zimmerman, A.R.; Green, S.J.; Uthandi, S.; Foster, J.S. Taxa-specific changes in soil microbial community composition induced by pyrogenic carbon amendments. Soil Biol. Biochem. 2011, 43, 385–392. [Google Scholar] [CrossRef]
- Carson, J.K.; Rooney, D.; Gleeson, D.B.; Clipson, N. Altering the balance: The response of ecosystem function and community composition to future climate change in a mesocosm study. Soil Biol. Biochem. 2010, 42, 1873–1881. [Google Scholar] [CrossRef]
- An, H.; Zhang, L.; Tang, Y.; Luo, X.; Sun, T.; Li, Y.; Wang, Y.; Dai, J.; Fang, C. Skermanella xinjiangensis sp. nov., isolated from the desert of Xinjiang, China. Int. J. Syst. Evol. Microbiol. 2009, 59 Pt 6, 1531–1534. [Google Scholar] [CrossRef] [Green Version]
- Aghnatios, A.; Drancourt, M. Gemmata species: Planctomycetes of medical interest. Future Microbiol. 2016, 11, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Whang, K.S. Reyranellagraminifolii sp. nov., isolated from bamboo (Phyllostachys bambusoides) litter. Int. J. Syst. Evol. Microbiol. 2014, 64, 2503–2507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales, M.E.; Allegrini, M.; Iocoli, G.A.; Basualdo, J.; Villamil, M.B.; Zabaloy, M.C. Rhizospheric Microbiome Responses to Cover Crop Suppression Methods. Agronomy 2022, 12, 2246. [Google Scholar] [CrossRef]
- Kiers, E.T.; Duhamel, M.; Beesetty, Y.; Mensah, J.A.; Franken, O.; Verbruggen, E.; Fellbaum, C.R.; Kowalchuk, G.A.; Hart, M.M.; Bago, A.; et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 2011, 333, 880–882. [Google Scholar] [CrossRef] [Green Version]
- Hartnett, D.C.; Wilson, G.W.; Gibson, D.J. Mycorrhizal influence on intra-and interspecific neighbor interactions among co-occurring prairie grasses. J. Ecol. 1993, 81, 787–795. [Google Scholar] [CrossRef]
- Allen, E.B.; Allen, M.F. Influence of early summer and fall prescribed burning on mycorrhizae of Quercus spp. in southern California. Restor. Ecol. 2002, 10, 626–632. [Google Scholar]
- Johnson, N.C.; Wilson, G.W.T.; Bowker, M.A.; Wilson, J.A.; Miller, R.M. Resource limitation is a driver of local adaptation in mycorrhizal symbioses. Proc. Natl. Acad. Sci. USA 2010, 107, 2093–2098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pattinson, G.S.; Hammill, K.A.; Sutton, B.G.; McGee, P.A. Simulated fire reduces the density of arbuscular mycorrhizal fungi at the soil surface. Mycol. Res. 1999, 103, 491–496. [Google Scholar] [CrossRef]
- Ahmadi, J.; Farzam, M.; Lagzian, A. Investigating effects of a prescribed spring fire on symbiosis between mycorrhizal fungi and range plant species. J. Rangel. Sci. 2017, 7, 138–147. [Google Scholar]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: Boston, MA, USA, 2008. [Google Scholar]
- Gazol, A.; Zobel, M.; Cantero, J.J.; Davison, J.; Esler, K.J.; Jairus, T.; Opik, M.; Vasar, M.; Moora, M. Impact of alien pines on local arbuscular mycorrhizal fungal communities-evidence from two continents. FEMS Microbiol. Ecol. 2016, 92, fiw073. [Google Scholar] [CrossRef] [Green Version]
- Redecker, D.; Schüβler, A.; Stockinger, H.; Stürmer, S.; Morton, J.; Walker, C. An evidence based consensus for the classification of arbuscular mycorrhizal fungi (Glomeromytoa). Mycorrhiza 2013, 23, 515–531. [Google Scholar] [CrossRef]
- Schalamuk, S.; Cabello, M. Arbuscular mycorrhizal fungal propagules from tillage and no-tillage systems: Possible effects on Glomeromycota diversity. Mycologia 2010, 102, 261–268. [Google Scholar] [CrossRef]
- Dove, N.C.; Hart, S.C. Fire reduces fungal species richness and in situ mycorrhizal colonization: A meta-analysis. Fire Ecol. 2017, 13, 37–65. [Google Scholar] [CrossRef]
- Shen, J.; Chen, C.; Lewis, T. Long term repeated fire disturbance alters soil bacterial diversity but not the abundance in an Australian wet sclerophyll forest. Sci. Rep. 2016, 6, 19639. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]




| Fire Treatment | Plot | Last Burnt | Period since Last Fire | Fires since Start of Trial |
|---|---|---|---|---|
| Annual autumn burn | 5 | May 2014 | 9 months | 34 |
| Annual spring burn | 12 | October 2014 | 4 months | 34 |
| Biennial autumn burn | 1 | May 2014 | 9 months | 18 |
| Biennial spring burn | 2 | October 2014 | 4 months | 18 |
| Five-year rotation burn | 6 | October 2012 | 27 months | 8 |
| No-burn treatments | 11 | 2007 (unplanned fire) | ±180 months | 2 |
| Soil Characteristic | Control | Five-Year | Biennial Autumn | Biennial Spring | Annual Autumn | Annual Spring |
|---|---|---|---|---|---|---|
| Ca (mg/kg) | 228.5 | 183.0 | 330.0 | 245.0 | 303.5 | 224.5 |
| Mg (mg/kg) | 85.0 | 77.5 | 103.5 | 86.0 | 97.0 | 78.5 |
| K (mg/kg) | 216.5 | 222.5 | 240.5 | 261.0 | 233.0 | 290.0 |
| Na (mg/kg) | 27.5 | 43.0 | 45.0 | 35.5 | 51.5 | 36.5 |
| P (mg/kg) | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| pH (H20) | 5.10 | 5.10 | 5.16 | 5.15 | 5.13 | 5.16 |
| pH (KCl) | 4.38 | 4.34 | 4.40 | 4.43 | 4.37 | 4.42 |
| EC (mS/m) | 14 | 10 | 12 | 12 | 12 | 13 |
| % Carbon (Walkley Black) | 11.17 | 13.32 | 11.56 | 14.10 | 10.97 | 12.93 |
| % Nitrogen (LECO) | 0.6 | 0.85 | 0.68 | 0.76 | 0.65 | 0.77 |
| Site | % Mycorrhizal Colonisation | AM Spores/100 g Soil |
|---|---|---|
| Control | 87.7 ± 6.1 | 198 ± 131.2 |
| Annual spring | 65.6 ± 21.3 | 575 ± 114.0 |
| Annual autumn | 67.9 ± 7.9 | 128 ± 72.1 |
| Biennial spring | 73.3 ± 13.4 | 94 ± 60.1 |
| Biennial autumn | 66.6 ± 5.6 | 289 ± 162.3 |
| Five-year rotation | 67.6 ± 4.5 | 287 ± 54.0 |
| Chi-square value | 11.595 | 12.135 |
| p value | 0.041 | 0.033 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gokul, J.K.; Matcher, G.; Dames, J.; Nkangala, K.; Gordijn, P.J.; Barker, N.P. Microbial Community Responses to Alterations in Historical Fire Regimes in Montane Grasslands. Diversity 2023, 15, 818. https://doi.org/10.3390/d15070818
Gokul JK, Matcher G, Dames J, Nkangala K, Gordijn PJ, Barker NP. Microbial Community Responses to Alterations in Historical Fire Regimes in Montane Grasslands. Diversity. 2023; 15(7):818. https://doi.org/10.3390/d15070818
Chicago/Turabian StyleGokul, Jarishma K., Gwynneth Matcher, Joanna Dames, Kuhle Nkangala, Paul J. Gordijn, and Nigel P. Barker. 2023. "Microbial Community Responses to Alterations in Historical Fire Regimes in Montane Grasslands" Diversity 15, no. 7: 818. https://doi.org/10.3390/d15070818