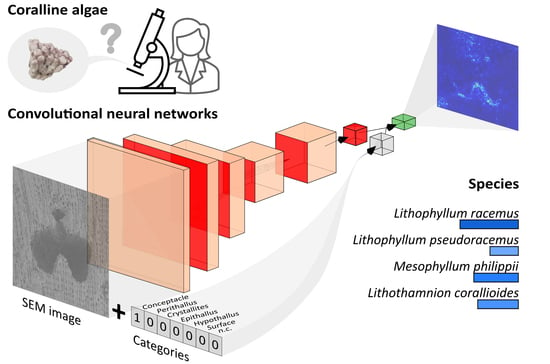
Deep Learning Applied to SEM Images for Supporting Marine Coralline Algae Classification
Abstract
:1. Introduction
- Discriminate L. pseudoracemus from all the other species (2 class-CNN);
- Classify the three genera (3 class-CNN);
- Classify the four species (4 class-CNN).
- We presented a new classification tool for coralline algae diagnosis, by applying a deep learning technique to SEM images for the automated identification of four species at different taxonomic levels;
- We developed and evaluated CNN-based classification models (open-sourced on GitHub as reported in the Data Availability Statement) against two baselines, namely a dummy classifier and a human-reported classification. Then, our model was tested in a practical scenario, to support the classification of two uncertain samples of coralline algae;
- We investigated and discussed the contribution of six main morphological categories, shown in the SEM images, to the classification task;
- We explored a set of explanation methods, which justify the class assignment of the proposed model by visually highlighting the contribution of portions of the processed SEM image.
2. Materials and Methods
2.1. Samples and Data Collection
2.2. Data Augmentation
2.3. Convolutional Neural Networks
- An input layer of fixed size 224 × 224 Red-Green-Blue image;
- A stack of convolutional layers, where the filters were used with a very small receptive field: 3 × 3;
- Five maxpooling layers (not all the convolutional layers are followed by max-pooling);
- A dense layer, whose input is the output of the previous maxpooling layer and concatenated with the one-hot-encoded categories, and;
- A softmax output layer.
- 2 class-CNN (L. pseudoracemus versus Others, i.e., all the other species), last dense layer of 128 neurons, output layer of two neurons;
- 3 class-CNN, at the genus level (Lithothamnion sp., Mesophyllum sp. and Lithophyllum sp.), last dense layer of 256 neurons, output layer of three neurons;
- 4 class-CNN, at the species level (L. corallioides, M. philippii, L. racemus and L. pseudoracemus), last dense layer of 64 neurons, output layer of four neurons.
2.4. Interpretability
2.5. Evaluation Protocol
3. Results
3.1. Internal Validation
Morphological Categories Analysis
3.2. External Test
Morphological Categories Analysis
3.3. Explanation
4. Discussion
- Conceptacles: ~250×, ~500×;
- Perithallus: ~1000×, ~2500×, ~5000×;
- Crystallites: ~10000×, ~20000×, ~30000×;
- Epithallus: ~1000×, ~2500×, ~5000×;
- Hypothallus: ~250×; ~1000×;
- Surface: ~1000×.
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


References
- Bracchi, V.A.; Basso, D.; Marchese, F.; Corselli, C.; Savini, A. Coralligenous morphotypes on subhorizontal substrate: A new categorization. Cont. Shelf Res. 2017, 144, 10–20. [Google Scholar] [CrossRef]
- Cebrián, E.; Ballesteros, E.; Canals, M. Shallow rocky bottom benthic assemblages as calcium carbonate producers in the Alboran Sea (Southwestern Mediterranean). Oceanol. Acta 2000, 23, 311–322. [Google Scholar] [CrossRef] [Green Version]
- Basso, D. Production carbonatée par les algues rouges calcaires et changement climatique global. Geodiversitas 2012, 34, 13–33. [Google Scholar] [CrossRef]
- Ballesteros, E. Mediterranean coralligenous assemblages: A synthesis of present knowledge. Oceanogr. Mar. Biol. 2006, 44, 123–195. [Google Scholar] [CrossRef]
- Basso, D.; Nalin, R.; Massari, F. Genesis and composition of the Pleistocene coralligène de plateau of the Cutro Terrace (Calabria, Southern Italy). Neues Jahrb. Geol. Palaontol. Abh. 2007, 244, 173–182. [Google Scholar] [CrossRef]
- Bracchi, V.A.; Nalin, R.; Basso, D. Morpho-structural heterogeneity of shallow-water coralligenous in a Pleistocene marine terrace (Le Castella, Italy). Palaeogeogr. Palaeoclimatol. Palaeoecol. 2016, 454, 101–112. [Google Scholar] [CrossRef]
- Basso, D.; Morbioli, C.; Corselli, C. Rhodolith facies evolution and burial as a response to Holocene transgression at the Pontian islands shelf break. Geol. Soc. Spec. Publ. 2006, 255, 23–34. [Google Scholar] [CrossRef]
- Bracchi, V.A.; Nalin, R.; Basso, D. Paleoecology and dynamics of coralline dominated facies during a Pleistocene transgressive-regressive cycle (Capo Colonna marine terrace, Southern Italy). Palaeogeogr. Palaeoclimatol. Palaeoecol. 2014, 414, 296–309. [Google Scholar] [CrossRef]
- Darrenougue, N.; de Deckker, P.; Eggins, S.; Payri, C. Sea-surface temperature reconstruction from trace elements variations of tropical coralline red algae. Quat. Sci. Rev. 2014, 93, 34–46. [Google Scholar] [CrossRef]
- Coletti, G.; Basso, D.; Corselli, C. Coralline algae as depth indicators in the Sommières Basin (Early Miocene, Southern France). Geobios 2018, 51, 15–30. [Google Scholar] [CrossRef]
- Basso, D. Study of living calcareous algae by a paleontological approach: The non-geniculate Corallinaceae (Rhodophyta) of the soft bottoms of the Tyrrhenian Sea (Western Mediterranean). The genera Phymatolithon Foslie and Mesophyllum Lemoine. Riv. Ital. Di Paleontol. Stratigr. 1995, 100, 575–596. [Google Scholar]
- Basso, D. Living calcareous algae by a paleontological approach: The genus Lithothamnion Heydrich Nom. Cons. from the soft bottoms of the Tyrrhenian Sea (Mediterranean). Riv. Ital. Di Paleontol. Stratigr. 1995, 101, 349–366. [Google Scholar] [CrossRef]
- Cabioch, J.; Mendoza, M.L. Mesophyllum Alternans (Foslie) Comb. Nov. (Corallinales, Rhodophyta), a Mediterraneo-Atlantic species, and new considerations on the Lithothamnion philippii Foslie complex. Phycologia 1998, 37, 208–221. [Google Scholar] [CrossRef]
- Adey, W.H.; Mcintyre, I.G. Crustose coralline algae: A re-evaluation in the geological sciences. Geol. Soc. Am. Bull. 1973, 84, 883–904. [Google Scholar] [CrossRef]
- Garbary, J.D. An introduction to the Scanning Electron Microscopy of red algae. In Modern Approaches to the Taxonomy of Red and Brown Algae; Academic Press: Cambridge, MA, USA, 1978; pp. 205–222. [Google Scholar]
- Caragnano, A.; Rodondi, G.; Basso, D.; Peña, V.; Gall, L.; Rindi, F. Circumscription of Lithophyllum racemus (Corallinales, Rhodophyta) from the Western Mediterranean Sea reveals the species Lithophyllum pseudoracemus sp. nov. Phycologia 2020, 59, 584–597. [Google Scholar] [CrossRef]
- Pezzolesi, L.; Peña, V.; le Gall, L.; Gabrielson, P.W.; Kaleb, S.; Hughey, J.R.; Rodondi, G.; Hernandez-Kantun, J.J.; Falace, A.; Basso, D.; et al. Mediterranean Lithophyllum stictiforme (Corallinales, Rhodophyta) is a genetically diverse species complex: Implications for species circumscription, biogeography and conservation of coralligenous habitats. J. Phycol. 2019, 55, 473–492. [Google Scholar] [CrossRef]
- De Jode, A.; David, R.; Haguenauer, A.; Cahill, A.E.; Erga, Z.; Guillemain, D.; Sartoretto, S.; Rocher, C.; Selva, M.; le Gall, L.; et al. From seascape ecology to population genomics and back. Spatial and ecological differentiation among cryptic species of the red algae Lithophyllum stictiforme/L. cabiochiae, main bioconstructors of coralligenous habitats. Mol. Phylogenetics Evol. 2019, 137, 104–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rösler, A.; Perfectti, F.; Peña, V.; Braga, J.C. Phylogenetic relationships of Corallinaceae (Corallinales, Rhodophyta): Taxonomic implications for reef-building corallines. J. Phycol. 2016, 52, 412–431. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Baba, M.; Suda, S. Revision of the Mastophoroideae (Corallinales, Rhodophyta) and polyphyly in nongeniculate species widely distributed on Pacific coral reefs. J. Phycol. 2011, 47, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Auer, G.; Piller, W.E. Nanocrystals as phenotypic expression of genotypes-An example in coralline red algae. Sci. Adv. 2020, 6, eaay2126. [Google Scholar] [CrossRef] [Green Version]
- Basso, D.; Fravega, P.; Vannucci, G. Fossil and living corallinaceans related to the Mediterranean endemic species Lithophyllum racemus (Lamarck) Foslie. Facies 1996, 35, 275–292. [Google Scholar] [CrossRef]
- Basso, D.; Babbini, L.; Kaleb, S.; Bracchi, V.A.; Falace, A. Monitoring deep Mediterranean rhodolith beds. Aquat. Conserv. Mar. Freshw. Ecosyst. 2016, 26, 549–561. [Google Scholar] [CrossRef] [Green Version]
- Basso, D. Deep rhodolith distribution in the Pontian islands, Italy: A model for the paleoecology of a temperate sea. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1998, 137, 173–187. [Google Scholar] [CrossRef]
- Foster, M.S. Rhodoliths: Between rocks and soft places. J. Phycol. 2001, 37, 659–667. [Google Scholar] [CrossRef]
- Basso, D.; Babbini, L.; Ramos-Esplá, A.A.; Salomidi, M. Mediterranean rhodolith beds. Coast. Res. Libr. 2017, 15, 281–298. [Google Scholar] [CrossRef]
- Carro, B.; Lopez, L.; Peña, V.; Bárbara, I.; Barreiro, R. DNA barcoding allows the accurate assessment of European maerl diversity: A proof-of-concept study. Phytotaxa 2014, 190, 176–189. [Google Scholar] [CrossRef] [Green Version]
- Potin, P.; Floc’h, J.Y.; Augris, C.; Cabioch, J. Annual growth rate of the calcareous red alga Lithothamnion corallioides (Corallinales, Rhodophyta) in the Bay of Brest, France. Hydrobiologia 1990, 204, 263–267. [Google Scholar] [CrossRef]
- Savini, A.; Basso, D.; Bracchi, V.A.; Corselli, C.; Pennetta, M. Cartographie du maërl et quantification de la production carbonatée sur les terrasses sous-marines au large de la péninsule du Cilento (Mer Tyrrhénienne, Italie). Geodiversitas 2012, 34, 77–98. [Google Scholar] [CrossRef]
- Martin, S.; Castets, M.D.; Clavier, J. Primary production, respiration and calcification of the temperate free-living coralline alga Lithothamnion corallioides. Aquat. Bot. 2006, 85, 121–128. [Google Scholar] [CrossRef]
- Irvine, L.M.; Chamberlain, Y. Seaweeds of the British isles: Volume 1 Rhodophyta. Part 2B Corallinales, Hildenbrandiales; Natural History Museum: London, UK, 1994; 284p. [Google Scholar]
- Peña, V.; de Clerck, O.; Afonso-Carrillo, J.; Ballesteros, E.; Bárbara, I.; Barreiro, R.; le Gall, L. An integrative systematic approach to species diversity and distribution in the genus Mesophyllum (Corallinales, Rhodophyta) in Atlantic and Mediterranean Europe. Eur. J. Phycol. 2015, 50, 20–36. [Google Scholar] [CrossRef]
- Park, J.-H.; Hwang, K.-B.; Park, H.-M.; Choi, Y.-K. Application of CNN for fish species classification. J. Korea Inst. Inf. Commun. Eng. 2019, 23, 39–46. [Google Scholar] [CrossRef]
- Rathi, D.; Jain, S.; Indu, S. Underwater fish species classification using Convolutional Neural Network and Deep Learning. In Proceedings of the 9th International Conference on Advances in Pattern Recognition 2018, Bangalore, India, 23–27 December 2017; pp. 344–349. [Google Scholar] [CrossRef] [Green Version]
- Kiel, S. Assessing bivalve phylogeny using Deep Learning and computer vision approaches. bioRxiv 2021. [Google Scholar] [CrossRef]
- De Lima, R.P.; Welch, K.F.; Barrick, J.E.; Marfurt, K.J.; Burkhalter, R.; Cassel, M.; Soreghan, G.S. Convolutional Neural Networks as an aid to biostratigraphy and micropaleontology: A test on Late Paleozoic microfossils. Palaios 2020, 35, 391–402. [Google Scholar] [CrossRef]
- Liu, X.; Song, H. Automatic identification of fossils and abiotic grains during carbonate microfacies analysis using Deep Convolutional Neural Networks. Sediment. Geol. 2020, 410, 105790. [Google Scholar] [CrossRef]
- Modarres, M.H.; Aversa, R.; Cozzini, S.; Ciancio, R.; Leto, A.; Brandino, G.P. Neural Network for nanoscience Scanning Electron Microscope image recognition. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Adey, W.H.; McKibbin, D.L. Studies on the maerl species Phymatolithon calcareum (Pallas) nov. comb. and Lithothamnium coralloides Crouan in the Ria de Vigo. Bot. Mar. 1970, 13, 100–106. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Hecht-Nielsen, R. Theory of the Backpropagation Neural Network. Int. Jt. Conf. Neural Netw. 1989, 1, 593–605. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, N.; Yang, W.; Wang, S.; Cui, Z.; Chen, X.; Chen, L. A review of image recognition with Deep Convolutional Neural Network. In Intelligent Computing Theories and Application; Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2017; Volume 10361 LNCS, pp. 69–80. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, K.; Liu, X.; Long, E.; Jiang, J.; An, Y.; Zhang, J.; Liu, Z.; Lin, Z.; Li, X.; et al. Comparative analysis of image classification methods for automatic diagnosis of ophthalmic images. Sci. Rep. 2017, 7, 41545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; 800p. [Google Scholar]
- Abdalla, A.; Cen, H.; Wan, L.; Rashid, R.; Weng, H.; Zhou, W.; He, Y. Fine-Tuning Convolutional Neural Network with Transfer Learning for semantic segmentation of ground-level oilseed rape images in a field with high weed pressure. Comput. Electron. Agric. 2019, 167, 105091. [Google Scholar] [CrossRef]
- Deng, Y.; Lu, L.; Aponte, L.; Angelidi, A.M.; Novak, V.; Karniadakis, G.E.; Mantzoros, C.S. Deep Transfer Learning and Data Augmentation improve glucose levels prediction in type 2 diabetes patients. NPJ Digit. Med. 2021, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F. ImageNet: A large-scale hierarchical image database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef] [Green Version]
- Agarap, A.F. Deep Learning Using Rectified Linear Units (ReLU). arXiv 2018, arXiv:1803.08375. [Google Scholar]
- Tan, P.; Steinbach, M.; Karpatne, A.; Kumar, V. Introduction to Data Mining, 2nd ed.; Addison-Wesley: Boston, MA, USA, 2018; 839p. [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside Convolutional Networks: Visualising image classification models and Saliency Maps. arXiv 2013, arXiv:1312.6034. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should I trust you?” Explaining the predictions of any classifier. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–16 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; Volume 13–17, pp. 1135–1144. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from Deep Networks via gradient-based localization. arXiv 2016, arXiv:1611.07450. [Google Scholar] [CrossRef] [Green Version]
- Chollet, F. Keras, GitHub. Available online: https://github.com/fchollet/keras (accessed on 19 November 2021).
- Kotikalapudi, R.; Contributors. Keras-Vis, GitHub. Available online: https://github.com/raghakot/keras-vis (accessed on 19 November 2021).
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-scale Machine Learning on heterogeneous distributed systems. arXiv 2015, arXiv:1603.04467. [Google Scholar]
- Hsiang, A.Y.; Brombacher, A.; Rillo, M.C.; Mleneck-Vautravers, M.J.; Conn, S.; Lordsmith, S.; Jentzen, A.; Henehan, M.J.; Metcalfe, B.; Fenton, I.S.; et al. Endless Forams: >34,000 modern planktonic foraminiferal images for taxonomic training and automated species recognition using Convolutional Neural Networks. Paleoceanogr. Paleoclimatology 2019, 34, 1157–1177. [Google Scholar] [CrossRef] [Green Version]
- Mitra, R.; Marchitto, T.M.; Ge, Q.; Zhong, B.; Kanakiya, B.; Cook, M.S.; Fehrenbacher, J.S.; Ortiz, J.D.; Tripati, A.; Lobaton, E. Automated species-level identification of planktic foraminifera using Convolutional Neural Networks, with comparison to human performance. Mar. Micropaleontol. 2019, 147, 16–24. [Google Scholar] [CrossRef]
- Flajs, G. Skeletal structures of some calcifying algae. In Fossil Algae; Springer: Berlin/Heidelberg, Germany, 1977; pp. 225–231. [Google Scholar] [CrossRef]
- Bracchi, V.A.; Piazza, G.; Basso, D. A stable ultrastructural pattern despite variable cell size in Lithothamnion corallioides. Biogeosciences 2021, 18, 6061–6076. [Google Scholar] [CrossRef]
- Nash, M.C.; Adey, W. Multiple phases of Mg-calcite in crustose coralline algae suggest caution for temperature proxy and ocean acidification assessment: Lessons from the ultrastructure and biomineralization in Phymatolithon (Rhodophyta, Corallinales). J. Phycol. 2017, 53, 970–984. [Google Scholar] [CrossRef] [PubMed]





| Sample | Species | Sampling Site (Latitude, Longitude) | Images (n) |
|---|---|---|---|
| iv1 | Lithothamnion corallioides | Villasimius, Sardinia (Italy) (39°08′32″ N, 9°31′14″ E) | 24 |
| iv2 | Morlaix, Brittany (France) (48°34′42″ N, 3°49′36″ W) | 25 | |
| iv3 | Mesophyllum philippii | Portofino, Liguria (Italy) (44°17′56″ N, 9°13′08″ E) | 11 |
| iv4 | Capraia, Tuscany (Italy) (43°01′04″ N, 9°46′26″ E) | 14 | |
| iv5 | Cavoli Island, Sardinia (Italy) (39°05′20″ N, 9°32′33″ E) | 16 | |
| iv6 | Lithophyllum racemus | Pontian Islands (Italy) (40°54′47″ N, 12°52′58″ E) | 24 |
| iv7 | Capri, Gulf of Naples (Italy) (40°34′08″ N, 14°13′32″ E) | 43 | |
| iv8 | Lithophyllum pseudoracemus | Pontian Islands (Italy) (40°11′43″ N, 12°53′07″ E) | 9 |
| iv9 | Villasimius, Sardinia (Italy) (39°08′32″ N, 9°31′14″ E) | 48 | |
| DB865 | Lithophyllum cf. racemus | Santa Catarina, Rovinj (Croatia) (45°04′32″ N, 13°37′38″ E) | 28 |
| DB866 | Torre dell’Orso, Puglia (Italy) (40°14′00″ N, 18°28′00″ E) | 13 |
| 2 classes | 3 classes | 4 classes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Class Recall | Class Recall | Class Recall | |||||||||
| CNN | HC | DC | CNN | HC | DC | CNN | HC | DC | |||
| L. pseudoracemus | 0.61 | 0.21 | 0.27 | Lithothamnion sp. | 0.55 | 0.73 | 0.23 | L. corallioides | 0.57 | 0.73 | 0.23 |
| Others | 0.62 | 0.92 | 0.73 | Mesophyllum sp. | 0.56 | 0.41 | 0.19 | M. philippii | 0.49 | 0.41 | 0.19 |
| Lithophyllum sp. | 0.69 | 0.42 | 0.58 | L. racemus | 0.46 | 0.33 | 0.31 | ||||
| L. pseudoracemus | 0.37 | 0.21 | 0.27 | ||||||||
| Global accuracy | 0.61 | 0.73 | 0.62 | 0.64 | 0.49 | 0.43 | 0.48 | 0.40 | 0.27 | ||
| 2 class-CNN | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. pseudoracemus | Others | |||||||||||||
| Category | Images (n) | Class Recall | Images (n) | Class Recall | ||||||||||
| conceptacles | 1 | 1.00 | 9 | 0.56 | ||||||||||
| perithallus | 7 | 0.14 | 32 | 0.53 | ||||||||||
| crystallites | 27 | 0.70 | 67 | 0.61 | ||||||||||
| epithallus | 10 | 0.70 | 24 | 0.71 | ||||||||||
| hypothallus | 0 | - | 3 | 0.67 | ||||||||||
| surface | 5 | 0.60 | 7 | 0.57 | ||||||||||
| n.c. | 2 | 1.00 | 4 | 0.75 | ||||||||||
| shared | 5 | 0.40 | 11 | 0.73 | ||||||||||
| 3 class-CNN | ||||||||||||||
| Lithothamnion sp. | Mesophyllum sp. | Lithophyllum sp. | ||||||||||||
| Category | Images (n) | Class Recall | Images (n) | Class Recall | Images (n) | Class Recall | ||||||||
| conceptacles | 0 | - | 4 | 0.75 | 6 | 0.67 | ||||||||
| perithallus | 20 | 0.65 | 3 | 0.33 | 16 | 0.44 | ||||||||
| crystallites | 14 | 0.57 | 13 | 0.38 | 67 | 0.85 | ||||||||
| epithallus | 12 | 0.42 | 5 | 0.20 | 17 | 0.47 | ||||||||
| hypothallus | 0 | - | 3 | 0.67 | 0 | - | ||||||||
| surface | 2 | 0.50 | 4 | 0.50 | 6 | 0.17 | ||||||||
| n.c. | 0 | - | 1 | 1.00 | 5 | 0.80 | ||||||||
| shared | 1 | 0.00 | 8 | 1.00 | 7 | 0.57 | ||||||||
| 4 class-CNN | ||||||||||||||
| L. corallioides | M. philippii | L. racemus | L. pseudoracemus | |||||||||||
| Category | Images (n) | Class Recall | Images (n) | Class Recall | Images (n) | Class Recall | Images (n) | Class Recall | ||||||
| conceptacles | 0 | - | 4 | 1.00 | 5 | 0.80 | 1 | 1.00 | ||||||
| perithallus | 20 | 0.60 | 3 | 0.33 | 9 | 0.22 | 7 | 0.00 | ||||||
| crystallites | 14 | 0.71 | 13 | 0.00 | 40 | 0.48 | 27 | 0.37 | ||||||
| epithallus | 12 | 0.50 | 5 | 0.20 | 7 | 0.57 | 10 | 0.70 | ||||||
| hypothallus | 0 | - | 3 | 0.67 | 0 | - | 0 | - | ||||||
| surface | 2 | 0.00 | 4 | 0.75 | 1 | 0.00 | 5 | 0.40 | ||||||
| n.c. | 0 | - | 1 | 1.00 | 3 | 0.33 | 2 | 0.00 | ||||||
| shared | 1 | 0.00 | 8 | 1.00 | 2 | 0.50 | 5 | 0.20 | ||||||
| 2 class-CNN | ||
|---|---|---|
| Sample DB865 | Sample DB866 | |
| L. pseudoracemus | 0.32 | 0.31 |
| Others | 0.68 | 0.69 |
| 3 class-CNN | ||
| Sample DB865 | Sample DB866 | |
| Lithothamnion sp. | 0.18 | 0.00 |
| Mesophyllum sp. | 0.00 | 0.08 |
| Lithophyllum sp. | 0.82 | 0.92 |
| 4 class-CNN | ||
| Sample DB865 | Sample DB866 | |
| L. corallioides | 0.18 | 0.08 |
| M. philippii | 0.00 | 0.08 |
| L. racemus | 0.50 | 0.54 |
| L. pseudoracemus | 0.32 | 0.30 |
| DB865 | 2 class-CNN | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Category | Images (n) | L. pseudoracemus | Others | |||||||
| conceptacles | 1 | 0.00 | 1.00 | |||||||
| perithallus | 3 | 0.67 | 0.33 | |||||||
| crystallites | 12 | 0.17 | 0.83 | |||||||
| epithallus | 5 | 0.20 | 0.80 | |||||||
| hypothallus | 0 | - | - | |||||||
| 1 surface | 2 | 0.50 | 0.50 | |||||||
| n.c. | 2 | 0.50 | 0.50 | |||||||
| shared | 3 | 0.67 | 0.33 | |||||||
| 3 class-CNN | ||||||||||
| Category | Images (n) | Lithothamnion sp. | Mesophyllum sp. | Lithophyllum sp. | ||||||
| conceptacles | 1 | 1.00 | 0.00 | 0.00 | ||||||
| perithallus | 3 | 0.33 | 0.00 | 0.67 | ||||||
| crystallites | 12 | 0.00 | 0.00 | 1.00 | ||||||
| epithallus | 5 | 0.40 | 0.00 | 0.60 | ||||||
| 1 surface | 2 | 0.50 | 0.00 | 0.50 | ||||||
| n.c. | 2 | 0.00 | 0.00 | 1.00 | ||||||
| shared | 0.00 | 0.00 | 1.00 | |||||||
| 4 class-CNN | ||||||||||
| Category | Images (n) | L. corallioides | M. philippii | L. racemus | L. pseudoracemus | |||||
| conceptacles | 1 | 0.00 | 0.00 | 1.00 | 0.00 | |||||
| perithallus | 3 | 0.33 | 0.00 | 0.00 | 0.67 | |||||
| crystallites | 12 | 0.00 | 0.00 | 0.83 | 0.17 | |||||
| epithallus | 5 | 0.80 | 0.00 | 0.20 | 0.00 | |||||
| 1 surface | 2 | 0.00 | 0.00 | 0.50 | 0.50 | |||||
| n.c. | 2 | 0.00 | 0.00 | 0.50 | 0.50 | |||||
| shared | 3 | 0.00 | 0.00 | 0.00 | 1.00 | |||||
| DB866 | 2 class-CNN | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Category | Images (n) | L. pseudoracemus | Others | |||||||
| conceptacles | 2 | 0.00 | 1.00 | |||||||
| perithallus | 2 | 1.00 | 0.00 | |||||||
| crystallites | 3 | 0.33 | 0.67 | |||||||
| epithallus | 1 | 0.00 | 1.00 | |||||||
| hypothallus | 0 | - | - | |||||||
| 1 surface | 1 | 1.00 | 0.00 | |||||||
| n.c. | 2 | 0.00 | 1.00 | |||||||
| shared | 2 | 0.00 | 1.00 | |||||||
| 3 class-CNN | ||||||||||
| Category | Images (n) | Lithothamnion sp. | Mesophyllum sp. | Lithophyllum sp. | ||||||
| conceptacles | 2 | 0.00 | 0.00 | 1.00 | ||||||
| perithallus | 2 | 0.00 | 0.00 | 1.00 | ||||||
| crystallites | 3 | 0.00 | 0.00 | 1.00 | ||||||
| epithallus | 1 | 0.00 | 0.00 | 1.00 | ||||||
| 1 surface | 1 | 0.00 | 0.00 | 1.00 | ||||||
| n.c. | 2 | 0.00 | 0.00 | 1.00 | ||||||
| shared | 2 | 0.00 | 0.50 | 0.50 | ||||||
| 4 class-CNN | ||||||||||
| Category | Images (n) | L. corallioides | M. philippii | L. racemus | L. pseudoracemus | |||||
| conceptacles | 2 | 0.00 | 0.00 | 1.00 | 0.00 | |||||
| perithallus | 2 | 0.00 | 0.00 | 0.00 | 1.00 | |||||
| crystallites | 3 | 0.00 | 0.00 | 0.67 | 0.33 | |||||
| epithallus | 1 | 1.00 | 0.00 | 0.00 | 0.00 | |||||
| 1 surface | 1 | 0.00 | 0.00 | 0.00 | 1.00 | |||||
| n.c. | 2 | 0.00 | 0.00 | 1.00 | 0.00 | |||||
| shared | 2 | 0.00 | 0.50 | 0.50 | 0.00 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piazza, G.; Valsecchi, C.; Sottocornola, G. Deep Learning Applied to SEM Images for Supporting Marine Coralline Algae Classification. Diversity 2021, 13, 640. https://doi.org/10.3390/d13120640
Piazza G, Valsecchi C, Sottocornola G. Deep Learning Applied to SEM Images for Supporting Marine Coralline Algae Classification. Diversity. 2021; 13(12):640. https://doi.org/10.3390/d13120640
Chicago/Turabian StylePiazza, Giulia, Cecile Valsecchi, and Gabriele Sottocornola. 2021. "Deep Learning Applied to SEM Images for Supporting Marine Coralline Algae Classification" Diversity 13, no. 12: 640. https://doi.org/10.3390/d13120640