Prognostic Value of Gut Microbiome for Conversion from Mild Cognitive Impairment to Alzheimer’s Disease Dementia within 4 Years: Results from the AlzBiom Study
Abstract
:1. Introduction
2. Results
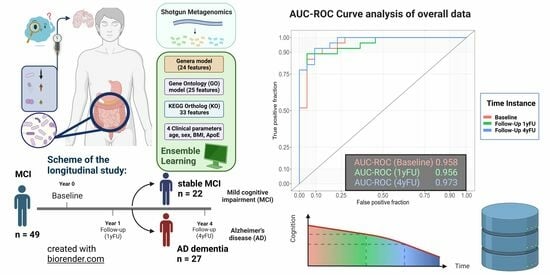
2.1. Discriminatory Ability of the Gut Microbiome between Stable MCI Patients and AD Dementia Converters
2.2. Longitudinal Development of Gut Microbiome
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Determination of Apolipoprotein E (ApoE) Genotype
4.3. Stool Collection, DNA Extraction and Shotgun Metagenomic Sequencing
4.4. Metagenomic Assembly
4.5. Taxonomic Profiling
4.6. Functional Profiling
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Albert, M.S.; DeKosky, S.T.; Dickson, D.; Dubois, B.; Feldman, H.H.; Fox, N.C.; Gamst, A.; Holtzman, D.M.; Jagust, W.J.; Petersen, R.C.; et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011, 7, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.J.; Shiri-Feshki, M. Rate of progression of mild cognitive impairment to dementia--meta-analysis of 41 robust inception cohort studies. Acta Psychiatr. Scand. 2009, 119, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Querfurth, H.W.; LaFerla, F.M. Alzheimer’s disease. N. Engl. J. Med. 2010, 362, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fang, L.; Chen, S.; Zhou, H.; Fan, Y.; Lin, L.; Li, J.; Xu, J.; Chen, Y.; Ma, Y.; et al. Gut Microbiome Alterations Precede Cerebral Amyloidosis and Microglial Pathology in a Mouse Model of Alzheimer’s Disease. BioMed Res. Int. 2020, 8456596. [Google Scholar] [CrossRef] [PubMed]
- Harach, T.; Marungruang, N.; Duthilleul, N.; Cheatham, V.; Mc Coy, K.D.; Frisoni, G.; Neher, J.J.; Fåk, F.; Jucker, M.; Lasser, T.; et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci. Rep. 2017, 7, 41802. [Google Scholar] [CrossRef] [PubMed]
- Dodiya, H.B.; Kuntz, T.; Shaik, S.M.; Baufeld, C.; Leibowitz, J.; Zhang, X.; Gottel, N.; Zhang, X.; Butovsky, O.; Gilbert, J.A.; et al. Sex-specific effects of microbiome perturbations on cerebral Aβ amyloidosis and microglia phenotypes. J. Exp. Med. 2019, 216, 1542–1560. [Google Scholar] [CrossRef]
- Dodiya, H.B.; Frith, M.; Sidebottom, A.; Cao, Y.; Koval, J.; Chang, E.; Sisodia, S.S. Synergistic depletion of gut microbial consortia, but not individual antibiotics, reduces amyloidosis in APPPS1-21 Alzheimer’s transgenic mice. Sci. Rep. 2020, 10, 8183. [Google Scholar] [CrossRef]
- Seo, D.O.; O’Donnell, D.; Jain, N.; Ulrich, J.D.; Herz, J.; Li, Y.; Lemieux, M.; Cheng, J.; Hu, H.; Serrano, J.R.; et al. ApoE isoform- and microbiota-dependent progression of neurodegeneration in a mouse model of tauopathy. Science 2023, 379, eadd1236. [Google Scholar] [CrossRef]
- Kim, M.S.; Kim, Y.; Choi, H.; Kim, W.; Park, S.; Lee, D.; Kim, D.K.; Kim, H.J.; Choi, H.; Hyun, D.W.; et al. Transfer of a healthy microbiota reduces amyloid and tau pathology in an Alzheimer’s disease animal model. Gut 2020, 69, 283–294. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef]
- Li, B.; He, Y.; Ma, J.; Huang, P.; Du, J.; Cao, L.; Wang, Y.; Xiao, Q.; Tang, H.; Chen, S. Mild cognitive impairment has similar alterations as Alzheimer’s disease in gut microbiota. Alzheimers Dement. 2019, 15, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wu, L.; Peng, G.; Han, Y.; Tang, R.; Ge, J.; Zhang, L.; Jia, L.; Yue, S.; Zhou, K.; et al. Altered microbiomes distinguish Alzheimer’s disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav. Immun. 2019, 80, 633–643. [Google Scholar] [CrossRef]
- Ling, Z.; Zhu, M.; Yan, X.; Cheng, Y.; Shao, L.; Liu, X.; Jiang, R.; Wu, S. Structural and Functional Dysbiosis of Fecal Microbiota in Chinese Patients with Alzheimer’s Disease. Front. Cell Dev. Biol. 2020, 8, 634069. [Google Scholar] [CrossRef] [PubMed]
- Laske, C.; Müller, S.; Preische, O.; Ruschil, V.; Munk, M.H.J.; Honold, I.; Peter, S.; Schoppmeier, U.; Willmann, M. Signature of Alzheimer’s Disease in Intestinal Microbiome: Results from the AlzBiom Study. Front. Neurosci. 2022, 16, 792996. [Google Scholar] [CrossRef]
- Ferreiro, A.L.; Choi, J.; Ryou, J.; Newcomer, E.P.; Thompson, R.; Bollinger, R.M.; Hall-Moore, C.; Ndao, I.M.; Sax, L.; Benzinger, T.L.S.; et al. Gut microbiome composition may be an indicator of preclinical Alzheimer’s disease. Sci. Transl. Med. 2023, 15, eabo2984. [Google Scholar] [CrossRef] [PubMed]
- Panagiotakos, D.B.; Pitsavos, C.; Stefanadis, C. Dietary patterns: A Mediterranean diet score and its relation to clinical and biological markers of cardiovascular disease risk. Nutr. Metab. Cardiovasc. Dis. 2006, 16, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Cerroni, R.; Pietrucci, D.; Teofani, A.; Chillemi, G.; Liguori, C.; Pierantozzi, M.; Unida, V.; Selmani, S.; Mercuri, N.B.; Stefani, A. Not just a Snapshot: An Italian Longitudinal Evaluation of Stability of Gut Microbiota Findings in Parkinson’s Disease. Brain Sci. 2022, 12, 739. [Google Scholar] [CrossRef] [PubMed]
- Emery, D.C.; Shoemark, D.K.; Batstone, T.E.; Waterfall, C.M.; Coghill, J.A.; Cerajewska, T.L.; Davies, M.; West, N.X.; Allen, S.J. 16S rRNA Next Generation Sequencing Analysis Shows Bacteria in Alzheimer’s Post-Mortem Brain. Front. Aging Neurosci. 2017, 9, 195. [Google Scholar] [CrossRef]
- Xi, J.; Ding, D.; Zhu, H.; Wang, R.; Su, F.; Wu, W.; Xiao, Z.; Liang, X.; Zhao, Q.; Hong, Z.; et al. Disturbed microbial ecology in Alzheimer’s disease: Evidence from the gut microbiota and fecal metabolome. BMC Microbiol. 2021, 21, 226. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Wang, J.; Chen, W.D.; Wang, Y.D. The Relationship Between Gut Microbiota and Inflammatory Diseases: The Role of Macrophages. Front. Microbiol. 2020, 11, 1065. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- McGeer, P.L.; McGeer, E.G. The inflammatory response system of brain: Implications for therapy of Alzheimer and other neurodegenerative diseases. Brain Res. Brain Res. Rev. 1995, 21, 195–218. [Google Scholar] [CrossRef]
- Wyss-Coray, T. Inflammation in Alzheimer disease: Driving force, bystander or beneficial response? Nat. Med. 2006, 12, 1005–1015. [Google Scholar] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Colombo, A.V.; Sadler, R.K.; Llovera, G.; Singh, V.; Roth, S.; Heindl, S.; Sebastian Monasor, L.; Verhoeven, A.; Peters, F.; Parhizkar, S.; et al. Microbiota-derived short chain fatty acids modulate microglia and promote Aβ plaque deposition. eLife 2021, 10, 59826. [Google Scholar] [CrossRef] [PubMed]
- Senizza, A.; Rocchetti, G.; Mosele, J.I.; Patrone, V.; Callegari, M.L.; Morelli, L.; Lucini, L. Lignans and Gut Microbiota: An Interplay Revealing Potential Health Implications. Molecules 2020, 25, 5709. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, C.; Li, R.; Liu, K.; Jin, G.; Ge, R.; Tang, F.; Cui, S. High-altitude Tibetan fermented milk ameliorated cognitive dysfunction by modified gut microbiota in Alzheimer’s disease transgenic mice. Food Funct. 2020, 11, 5308–5319. [Google Scholar] [CrossRef]
- Suhocki, P.V.; Ronald, J.S.; Diehl, A.M.E.; Murdoch, D.M.; Doraiswamy, P.M. Probing gut-brain links in Alzheimer’s disease with rifaximin. Alzheimers Dement. 2022, 8, e12225. [Google Scholar] [CrossRef]
- Liu, C.; Sun, D.; Zhu, J.; Liu, W. Two-Component Signal Transduction Systems: A Major Strategy for Connecting Input Stimuli to Biofilm Formation. Front. Microbiol. 2018, 9, 3279. [Google Scholar] [CrossRef]
- Erskine, E.; MacPhee, C.E.; Stanley-Wall, N.R. Functional Amyloid and Other Protein Fibers in the Biofilm Matrix. J. Mol. Biol. 2018, 430, 3642–3656. [Google Scholar] [CrossRef] [PubMed]
- Tükel, C.; Wilson, R.P.; Nishimori, J.H.; Pezeshki, M.; Chromy, B.A.; Bäumler, A.J. Responses to amyloids of microbial and host origin are mediated through toll-like receptor 2. Cell Host Microbe 2009, 6, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.L.; Bessho, S.; Grando, K.; Tükel, Ç. Microbiome or Infections: Amyloid-Containing Biofilms as a Trigger for Complex Human Diseases. Front. Immunol. 2021, 12, 638867. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.D.; Martins, F.; Wiltfang, J.; da Cruz, E.S.O.A.B.; Rebelo, S. ABC Transporters Are Key Players in Alzheimer’s Disease. J. Alzheimers Dis. 2018, 61, 463–485. [Google Scholar] [CrossRef] [PubMed]
- Conway, M.E. Alzheimer’s disease: Targeting the glutamatergic system. Biogerontology 2020, 21, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Morris, J.C. The Clinical Dementia Rating (CDR): Current version and scoring rules. Neurology 1993, 43, 2412–2414. [Google Scholar] [CrossRef]
- Morris, J.C. Clinical dementia rating: A reliable and valid diagnostic and staging measure for dementia of the Alzheimer type. Int. Psychogeriatr. 1997, 9 (Suppl. S1), 173–176; discussion 177–178. [Google Scholar] [CrossRef]
- Petersen, R.C. Mild cognitive impairment as a diagnostic entity. J. Intern. Med. 2004, 256, 183–194. [Google Scholar] [CrossRef]
- Negash, S.; Geda, Y.E.; Petersen, R.C. Neuropsychological characterization of mild cognitive impairment. Handb. Clin. Neurol. 2008, 88, 499–509. [Google Scholar]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Operto, G.; Molinuevo, J.L.; Cacciaglia, R.; Falcon, C.; Brugulat-Serrat, A.; Suárez-Calvet, M.; Grau-Rivera, O.; Bargalló, N.; Morán, S.; Esteller, M.; et al. Interactive effect of age and APOE-ε4 allele load on white matter myelin content in cognitively normal middle-aged subjects. Neuroimage Clin. 2019, 24, 101983. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.B.F.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; Ohara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. vegan: Community Ecology Package. 2022. Available online: https://github.com/vegandevs/vegan (accessed on 30 January 2024).
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife 2021, 10, 65088. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Riedel, B.C.; Thompson, P.M.; Brinton, R.D. Age, APOE and sex: Triad of risk of Alzheimer’s disease. J. Steroid Biochem. Mol. Biol. 2016, 160, 134–147. [Google Scholar] [CrossRef]
- Kang, S.Y.; Kim, Y.J.; Jang, W.; Son, K.Y.; Park, H.S.; Kim, Y.S. Body mass index trajectories and the risk for Alzheimer’s disease among older adults. Sci. Rep. 2021, 11, 3087. [Google Scholar] [CrossRef]
- De la Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. mSystems 2019, 4, 10–1128. [Google Scholar] [CrossRef]
- Haro, C.; Rangel-Zúñiga, O.A.; Alcalá-Díaz, J.F.; Gómez-Delgado, F.; Pérez-Martínez, P.; Delgado-Lista, J.; Quintana-Navarro, G.M.; Landa, B.B.; Navas-Cortés, J.A.; Tena-Sempere, M.; et al. Intestinal Microbiota Is Influenced by Gender and Body Mass Index. PLoS ONE 2016, 11, e0154090. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.T.; Corsini, S.; Kellingray, L.; Hegarty, C.; Le Gall, G.; Narbad, A.; Müller, M.; Tejera, N.; O’Toole, P.W.; Minihane, A.M.; et al. APOE genotype influences the gut microbiome structure and function in humans and mice: Relevance for Alzheimer’s disease pathophysiology. FASEB J. 2019, 33, 8221–8231. [Google Scholar] [CrossRef] [PubMed]
- Brunner, E.; Bathke, A.C.; Konietschke, F. Rank and Pseudo-Rank Procedures for Independent Observations in Factorial Designs; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Noguchi, K.; Gel, Y.R.; Brunner, E.; Konietschke, F. nparLD: An R Software Package for the Nonparametric Analysis of Longitudinal Data in Factorial Experiments. J. Stat. Softw. 2012, 50, 1–23. [Google Scholar] [CrossRef]
- Bischl, B.; Lang, M.; Kotthoff, L.; Schiffner, J.; Richter, J.; Studerus, E.; Casalicchio, G.; Jones, Z.M. mlr: Machine Learning in R. J. Mach. Learn. Res. 2016, 17, 5938–5942. [Google Scholar]



| Stable MCI | AD Dementia Converters | p-Value | |
|---|---|---|---|
| Number (n) | 22 | 27 | |
| Age, years, mean (SD) | 70.4 (6.9) | 72.9 (7.7) | 0.2492 |
| Gender (m/f) | 10/12 | 8/19 | 0.3723 |
| MMSE at baseline, mean (SD) | 26.9 (1.4) | 26.9 (1.8) | 0.9569 |
| MMSE at 1-year follow-up (1yFU), mean (SD) | 26.4 (1.8) | 24.4 (3.2) | 0.0108 |
| MMSE at 4-year follow-up (4yFU), mean (SD) | 25.9 (1.4) | 20.2 (4.3) | <0.0001 |
| GDS, mean (SD) | 2.0 (1.6) | 2.4 (1.8) | 0.4493 |
| Body mass index (BMI), mean (SD) | 25.1 (3.8) | 26.2 (4.7) | 0.3715 |
| ApoE (e4/e4 carriers/single e4 carriers/non-e4-carriers; n) | 1/6/6 | 0/5/10 | 0.3757 |
| Arterial hypertension (yes/no) | 6/16 | 14/13 | 0.1433 |
| Diabetes mellitus (yes/no) | 0/22 | 3/24 | 0.2423 |
| Rheumatoid arthritis (yes/no) | 1/21 | 0/27 | 0.4490 |
| NSAIDs (yes/no) | 6/16 | 8/19 | 0.8559 |
| Anticoagulants (yes/no) | 0/22 | 0/27 | n.a. |
| Antihypertensives (yes/no) | 6/16 | 13/14 | 0.1551 |
| Antidiabetics (yes/no) | 0/22 | 1/26 | 0.9853 |
| Statins (yes/no) | 4/18 | 7/20 | 0.7325 |
| Antidepressants (yes/no) | 2/20 | 7/20 | 0.1595 |
| AChE inhibitors (yes/no) | 1/21 | 4/23 | 0.3622 |
| Mediterranean diet score [16] | 32.0 (5.1) | 30.1 (5.2) | 0.1141 |
| Genus | Phylum | Genera Levels in AD Dementia Converters vs. Stable MCI Patients (↑ Increased, ↓ Decreased) |
|---|---|---|
| Clostridium | Bacillota | ↓ |
| Erysipelatoclostridium | Bacillota | ↑ *** |
| Mediterranea | Bacteroidota | ↓ |
| Lawsonibacter | Bacillota | ↓ |
| Sebaldella | Fusobacteriota | ↑ ** |
| Haloglycomyces | Actinomycetota | ↑ |
| Actinoalloteichus | Actinomycetota | ↑ |
| Arsenicicoccus | Actinomycetota | ↑ |
| Serinicoccus | Actinomycetota | ↑ |
| Knoellia | Actinomycetota | ↓ |
| Proteiniclasticum | Bacillota | ↑ |
| Alkaliflexus | Bacteroidota | ↓ * |
| Geofilum | Bacteroidota | ↓ * |
| Filimonas | Bacteroidota | ↑ ** |
| Terrimonas | Bacteroidota | ↓ |
| Pricia | Bacteroidota | ↓ |
| Lautropia | Pseudomonadota | ↓ |
| Acidiphilium | Pseudomonadota | ↓ |
| Komagataeibacter | Pseudomonadota | ↑ |
| Teredinibacter | Pseudomonadota | ↑ ** |
| Methyloglobulus | Pseudomonadota | ↑ |
| Oceanococcus | Pseudomonadota | ↑ |
| Persephonella | Aquificota | ↑ * |
| Kutzneria | Actinomycetota | ↓ |
| KO Label | Name | Pathways/Reaction Mechanisms | Relative Abundance of KO Values in AD Dementia Converters vs. Stable MCI Patients (↑ Increased, ↓ Decreased) |
|---|---|---|---|
| K02474 | UDP-N-acetyl-D-galactosamine dehydrogenase | Metabolism of amino sugar and nucleotide sugar Biosynthesis of O-Antigen nucleotide sugar Biosynthesis of nucleotide sugars | ↑ * |
| K17235 | Arabinosaccharide transport system permease protein | ATP-binding cassette (ABC) transporters | ↓ |
| K02405 | RNA polymerase sigma factor for flagellar operon FliA | Two-component system Biofilm formation of pseudomonas aeruginosa, escherichia coli and vibrio cholerae Flagellar assembly | ↓ |
| K07742 | Uncharacterized protein | Unclassified | ↓ ** |
| K02041 | Phosphonate transport system ATP-binding protein | ABC transporters | ↓ |
| K03826 | Putative acetyltransferase | Protein modification | ↓ * |
| K03706 | Transcriptional pleiotropic repressor | Senses the intracellular pool of branched-chain amino acids | ↓ |
| K01951 | Guanosine monophosphate (GMP) synthase | ATP + xanthosine 5’-phosphate + L-glutamine + H2O ⇌ AMP + diphosphate + GMP + L-glutamate Metabolism of purine, drugs and nucleotides | ↑ |
| K02065 | Phospholipid/cholesterol/gamma-HCH transport system ATP-binding protein | ABC transporters | ↑ |
| K00765 | ATP phosphoribosyltransferase | Histidine metabolism Metabolic pathways Biosynthesis of secondary metabolites and amino acids | ↑ |
| K09789 | Pimeloyl-[acyl-carrier protein] methyl ester esterase | Biotin metabolism Biosynthesis of cofactors | ↑ * |
| K02654 | Leader peptidase (prepilin peptidase)/N-methyltransferase | Formation of pseudopili | ↓ |
| K09766 | Uncharacterized protein | Unclassified | ↓ |
| K03408 | Purine-binding chemotaxis protein CheW | Two-component system Bacterial chemotaxis | ↓ |
| K03270 | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (KDO 8-P phosphatase) | Biosynthesis of lipopolysaccharides and nucleotide sugars Metabolic pathways | ↑ |
| K02396 | Flagellar hook-associated protein 1 | Flagellar assembly | ↓ |
| K01923 | Phosphoribosylaminoimidazole-succinocarboxamide synthase | Purine metabolism Biosynthesis of secondary metabolites | ↑ |
| K01736 | Chorismate synthase | Biosynthesis of phenylalanine, tyrosine, tryptophan, secondary metabolites and amino acids Metabolic pathways | ↑ |
| K18928 | L-lactate dehydrogenase complex protein LldE | Conversion of pyruvate to lactate and back | ↑ |
| K02387 | Flagellar basal-body rod protein FlgB | Flagellar assembly | ↓ |
| K00575 | Chemotaxis protein methyltransferase CheR | Two-component system Bacterial chemotaxis | ↓ |
| K07718 | Two-component system, sensor histidine kinase YesM | Two-component system | ↓ |
| K06378 | Stage II sporulation protein AA (anti-sigma F factor antagonist) | Regulation of DNA-templated transcription Cellular spore formation | ↓ * |
| K06023 | HPr kinase/phosphorylase | Catabolite repression in Gram-positive bacteria Phosphorylates HPr, a phosphocarrier protein of a sugar transport and phosphorylation system at a serine residue | ↓ |
| K05350 | Beta-glucosidase | Metabolism of cyanoamino acid, starch and sucrose Degradation of flavonoids Biosynthesis of various plant secondary metabolites | ↓ |
| K00338 | NADH-quinone oxidoreductase subunit I | Oxidative phosphorylation Metabolic pathways | ↑ * |
| K00281 | Glycine dehydrogenase | Metabolism of glycine, serine, threonine, glyoxylate, dicarboxylate, lipoic acid and carbon Biosynthesis of secondary metabolites | ↑ ** |
| K06143 | Inner membrane protein | Unclassified | ↑ * |
| K01308 | Gamma-D-glutamyl-meso-diaminopimelate peptidase | Endopeptidase | ↓ |
| K13652 | AraC family transcriptional regulator | Binds to the target DNA and regulates bacterial virulence by sensing small molecule inducers | ↑ * |
| K02398 | Negative regulator of flagellin synthesis FlgM | Two-component system Biofilm formation of pseudomonas aeruginosa and escherichia coli Flagellar assembly | ↓ |
| K07814 | Putative two-component system response regulator | Putative two-component system response regulation | ↓ * |
| K01277 | Dipeptidyl-peptidase III | Intracellular peptide catabolism | ↑ |
| GO Label | NAME/TERM | Definition/Reaction Mechanisms | Relative Abundance of GO Values in AD Dementia Converters vs. Stable MCI Patients (↑ Increased, ↓ Decreased) |
|---|---|---|---|
| GO.0050549 | Cyclohexyl-isocyanide hydratase activity | N-cyclohexylformamide + H+ = cyclohexyl isocyanide + H2O | ↑ ** |
| GO.0015716 | Organic phosphonate transport | Alkylphosphonate transport | ↑ * |
| GO.0047536 | 2-aminoadipate transaminase activity | 2-oxoglutarate + L-2-aminoadipate = 2-oxoadipate + L-glutamate | ↑ ** |
| GO.0006084 | Acetyl-CoA metabolic process | Key intermediate in lipid and terpenoid biosynthesis | ↓ ** |
| GO.0006694 | Steroid biosynthetic process | Formation of steroids | ↑ |
| GO.0009986 | Cell surface binding | Component of the cell wall and/or plasma membrane | ↓ * |
| GO.0016999 | Antibiotic metabolic process | Antibiotic metabolism | ↓ ** |
| GO.0003935 | GTP cyclohydrolase II activity | GTP + 3 H2O = 2,5-diamino-6-hydroxy-4-(5-phosphoribosylamino)-pyrimidine + diphosphate + formate + 3 H+ | ↓ * |
| GO.0004634 | Phosphopyruvate hydratase activity | 2-phospho-D-glycerate = phosphoenolpyruvate + H2O | ↓ |
| GO.0006399 | tRNA metabolic process | tRNA metabolism | ↑ |
| GO.0016854 | Racemase and epimerase activity | Configuration change in one or more chiral centers in a molecule | ↑ |
| GO.0046405 | Glycerol dehydratase activity | Glycerol = 3-hydroxypropanal + H2O | ↑ |
| GO.0052907 | 23S rRNA (adenine(1618)-N(6))-methyltransferase activity | S-adenosyl-L-methionine + adenine (1618) in 23S rRNA = S-adenosyl-L-homocysteine + rRNA containing N(6)-methyladenine (1618) in 23S rRNA | ↑ * |
| GO.0006974 | DNA damage response | Cellular response to DNA damage | ↑ * |
| GO.2000143 | Negative regulation of DNA-templated transcription initiation | Limitation of DNA-templated transcription initiation | ↑ * |
| GO.0002143 | tRNA wobble position uridine thiolation | Post-transcriptional thiolation at the C2 position of an uridine residue at position 34 in the anticodon of a tRNA | ↑ ** |
| GO.0019323 | Pentose catabolic process | Breakdown of a pentose, any monosaccharide with a chain of five carbon atoms in the molecule | ↑ * |
| GO.0046104 | Thymidine metabolic process | Thymidine metabolism | ↑ * |
| GO.0016041 | Glutamate synthase (ferredoxin) activity | 2 L-glutamate + 2 oxidized ferredoxin = L-glutamine + 2-oxoglutarate + 2 reduced ferredoxin + 2 H+ | ↓ |
| GO.0016776 | Phosphotransferase activity, phosphate group as acceptor | Transfer of a phosphorus-containing group from one compound (donor) to a phosphate group (acceptor) | ↑ * |
| GO.0033764 | Steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | Oxidation-reduction (redox) reaction in which a CH-OH group acts as a hydrogen or electron donor and reduces NAD+ or NADP, and in which one substrate is a sterol derivative | ↑ * |
| GO.0010133 | Proline catabolic process to glutamate | Proline degradation to glutamate | ↑ |
| GO.0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity | H2O + L-glutamate 5-semialdehyde + NAD+ = 2 H+ + L-glutamate + NADH | ↑ |
| GO.0004657 | Proline dehydrogenase activity | L-proline + acceptor = (S)-1-pyrroline-5-carboxylate + reduced acceptor (first of two enzymatic reactions in proline degradation to glutamate) | ↑ * |
| GO.0004775 | Succinate-CoA ligase (ADP-forming) activity | ATP + succinate + CoA = ADP + succinyl-CoA + phosphate | ↑ ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laske, C.; Müller, S.; Munk, M.H.J.; Honold, I.; Willmann, M.; Peter, S.; Schoppmeier, U. Prognostic Value of Gut Microbiome for Conversion from Mild Cognitive Impairment to Alzheimer’s Disease Dementia within 4 Years: Results from the AlzBiom Study. Int. J. Mol. Sci. 2024, 25, 1906. https://doi.org/10.3390/ijms25031906
Laske C, Müller S, Munk MHJ, Honold I, Willmann M, Peter S, Schoppmeier U. Prognostic Value of Gut Microbiome for Conversion from Mild Cognitive Impairment to Alzheimer’s Disease Dementia within 4 Years: Results from the AlzBiom Study. International Journal of Molecular Sciences. 2024; 25(3):1906. https://doi.org/10.3390/ijms25031906
Chicago/Turabian StyleLaske, Christoph, Stephan Müller, Matthias H. J. Munk, Iris Honold, Matthias Willmann, Silke Peter, and Ulrich Schoppmeier. 2024. "Prognostic Value of Gut Microbiome for Conversion from Mild Cognitive Impairment to Alzheimer’s Disease Dementia within 4 Years: Results from the AlzBiom Study" International Journal of Molecular Sciences 25, no. 3: 1906. https://doi.org/10.3390/ijms25031906