Applications and Advances of Multicellular Tumor Spheroids: Challenges in Their Development and Analysis
Abstract
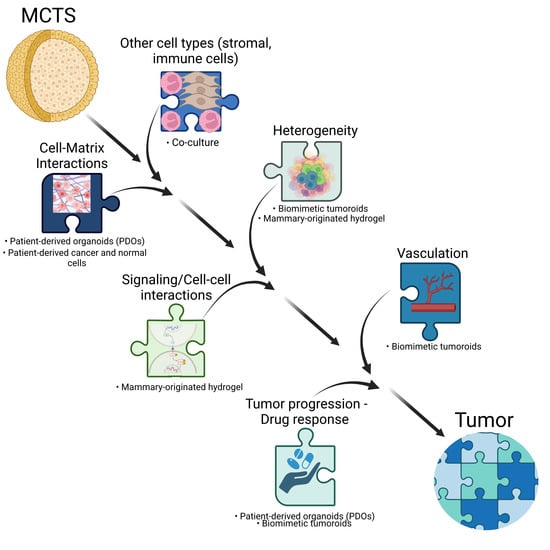
:1. Introduction
2. Methods of MCTS Formation
2.1. Scaffold-Free Systems
2.1.1. Liquid Overlay Technique
2.1.2. Hanging Drop Assay
2.1.3. Agitation-Based Methods
2.2. Scaffold-Based Systems
| Scaffold Type | Properties |
|---|---|
| Collagen [69,70,71] |
|
| Matrigel [45] |
|
| Fibrin [46,72,73] |
|
| Cellulose [49,74,75] |
|
| Starch [76,77] |
|
| Agarose [52,53] |
|
| Glycogen [55,78] |
|
| Hyaluronic acid (HA) [79,80,81] |
|
| Polypeptides [58,82] |
|
| Polyacrylamide (PA) [83,84] |
|
| Polyethylene glycol (PEG) [85] |
|
3. Applications of MCTSs in Anti-Cancer Studies
3.1. Cancer Cell Metabolism
3.2. Cell Cycle Research
4. Imaging and Analysis of MCTS
3D Cultures vs. In Vivo Tumors—What Is Missing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Kunz-Schughart, L.A.; Freyer, J.P.; Hofstaedter, F.; Ebner, R. The use of 3-D cultures for high-throughput screening: The multicellular spheroid model. J. Biomol. Screen 2004, 9, 273–285. [Google Scholar] [CrossRef] [Green Version]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Courau, T.; Bonnereau, J.; Chicoteau, J.; Bottois, H.; Remark, R.; Assante Miranda, L.; Toubert, A.; Blery, M.; Aparicio, T.; Allez, M.; et al. Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment. J. Immunother. Cancer 2019, 7, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyga, A.; Cheema, U.; Loizidou, M. 3D tumour models: Novel in vitro approaches to cancer studies. J. Cell Commun. Signal. 2011, 5, 239–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. 2017, 22, 456–472. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.L.; Jiang, Q.; Han, S.; Wu, Y.; Cui Tomshine, J.; Wang, D.; Gan, Y.; Zou, G.; Liang, X.J. Multicellular tumor spheroids as an in vivo-like tumor model for three-dimensional imaging of chemotherapeutic and nano material cellular penetration. Mol. Imaging 2012, 11, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Sabanayagam, C.R.; Harrington, D.A.; Farach-Carson, M.C.; Jia, X. A hydrogel-based tumor model for the evaluation of nanoparticle-based cancer therapeutics. Biomaterials 2014, 35, 3319–3330. [Google Scholar] [CrossRef] [Green Version]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef] [Green Version]
- Leung, B.M.; Lesher-Perez, S.C.; Matsuoka, T.; Moraes, C.; Takayama, S. Media additives to promote spheroid circularity and compactness in hanging drop platform. Biomater. Sci. 2015, 3, 336–344. [Google Scholar] [CrossRef] [Green Version]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef] [PubMed]
- Hirschhaeuser, F.; Menne, H.; Dittfeld, C.; West, J.; Mueller-Klieser, W.; Kunz-Schughart, L.A. Multicellular tumor spheroids: An underestimated tool is catching up again. J. Biotechnol. 2010, 148, 3–15. [Google Scholar] [CrossRef]
- Mollica, P.A.; Booth-Creech, E.N.; Reid, J.A.; Zamponi, M.; Sullivan, S.M.; Palmer, X.L.; Sachs, P.C.; Bruno, R.D. 3D bioprinted mammary organoids and tumoroids in human mammary derived ECM hydrogels. Acta Biomater. 2019, 95, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Magdeldin, T.; Lopez-Davila, V.; Pape, J.; Cameron, G.W.; Emberton, M.; Loizidou, M.; Cheema, U. Engineering a vascularised 3D in vitro model of cancer progression. Sci. Rep. 2017, 7, 44045. [Google Scholar] [CrossRef] [Green Version]
- Cabeza-Segura, M.; Garcia-Mico, B.; La Noce, M.; Nicoletti, G.F.; Conti, V.; Filippelli, A.; Fleitas, T.; Cervantes, A.; Castillo, J.; Papaccio, F. How organoids can improve personalized treatment in patients with gastro-esophageal tumors. Curr. Opin. Pharmacol. 2023, 69, 102348. [Google Scholar] [CrossRef] [PubMed]
- Nyga, A.; Stamati, K.; Redondo, P.A.; Azimi, T.; Feber, A.; Neves, J.B.; Hamoudi, R.; Presneau, N.; El Sheikh, S.; Tran, M.G.B.; et al. Renal tumouroids: Challenges of manufacturing 3D cultures from patient derived primary cells. J. Cell Commun. Signal. 2022, 16, 637–648. [Google Scholar] [CrossRef]
- Papaccio, F.; Cabeza-Segura, M.; Garcia-Mico, B.; Tarazona, N.; Roda, D.; Castillo, J.; Cervantes, A. Will Organoids Fill the Gap towards Functional Precision Medicine? J. Pers. Med. 2022, 12, 1939. [Google Scholar] [CrossRef]
- Lee, G.Y.; Kenny, P.A.; Lee, E.H.; Bissell, M.J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 2007, 4, 359–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokhari, M.; Carnachan, R.J.; Cameron, N.R.; Przyborski, S.A. Culture of HepG2 liver cells on three dimensional polystyrene scaffolds enhances cell structure and function during toxicological challenge. J. Anat. 2007, 211, 567–576. [Google Scholar] [CrossRef]
- Yamada, K.M.; Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanoni, M.; Pignatta, S.; Arienti, C.; Bonafe, M.; Tesei, A. Anticancer drug discovery using multicellular tumor spheroid models. Expert Opin. Drug Discov. 2019, 14, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, M.; Piccinini, F.; Arienti, C.; Zamagni, A.; Santi, S.; Polico, R.; Bevilacqua, A.; Tesei, A. 3D tumor spheroid models for in vitro therapeutic screening: A systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016, 6, 19103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivascu, A.; Kubbies, M. Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J. Biomol. Screen 2006, 11, 922–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedrich, J.; Seidel, C.; Ebner, R.; Kunz-Schughart, L.A. Spheroid-based drug screen: Considerations and practical approach. Nat. Protoc. 2009, 4, 309–324. [Google Scholar] [CrossRef]
- Carvalho, E.O.; Ribeiro, C.; Correia, D.M.; Botelho, G.; Lanceros-Mendez, S. Biodegradable Hydrogels Loaded with Magnetically Responsive Microspheres as 2D and 3D Scaffolds. Nanomaterials 2020, 10, 2421. [Google Scholar] [CrossRef]
- Metzger, W.; Sossong, D.; Bachle, A.; Putz, N.; Wennemuth, G.; Pohlemann, T.; Oberringer, M. The liquid overlay technique is the key to formation of co-culture spheroids consisting of primary osteoblasts, fibroblasts and endothelial cells. Cytotherapy 2011, 13, 1000–1012. [Google Scholar] [CrossRef]
- Lin, R.Z.; Chang, H.Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008, 3, 1172–1184. [Google Scholar] [CrossRef]
- Thoma, C.R.; Zimmermann, M.; Agarkova, I.; Kelm, J.M.; Krek, W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 2014, 69–70, 29–41. [Google Scholar] [CrossRef]
- Ryu, N.E.; Lee, S.H.; Park, H. Spheroid Culture System Methods and Applications for Mesenchymal Stem Cells. Cells 2019, 8, 1620. [Google Scholar] [CrossRef] [Green Version]
- Bartosh, T.J.; Ylostalo, J.H. Preparation of anti-inflammatory mesenchymal stem/precursor cells (MSCs) through sphere formation using hanging-drop culture technique. Curr. Protoc. Stem Cell Biol. 2014, 28, 2B.6.1–2B.6.23. [Google Scholar] [CrossRef] [Green Version]
- Kelm, J.M.; Timmins, N.E.; Brown, C.J.; Fussenegger, M.; Nielsen, L.K. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 2003, 83, 173–180. [Google Scholar] [CrossRef]
- Bialkowska, K.; Komorowski, P.; Bryszewska, M.; Milowska, K. Spheroids as a Type of Three-Dimensional Cell Cultures-Examples of Methods of Preparation and the Most Important Application. Int. J. Mol. Sci. 2020, 21, 6225. [Google Scholar] [CrossRef]
- Kim, J.B. Three-dimensional tissue culture models in cancer biology. Semin Cancer Biol 2005, 15, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, T.J.; Prewett, T.L.; Wolf, D.A.; Spaulding, G.F. Reduced shear stress: A major component in the ability of mammalian tissues to form three-dimensional assemblies in simulated microgravity. J. Cell Biochem. 1993, 51, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Rauh, J.; Milan, F.; Gunther, K.P.; Stiehler, M. Bioreactor systems for bone tissue engineering. Tissue Eng. Part B Rev. 2011, 17, 263–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingram, M.; Techy, G.B.; Saroufeem, R.; Yazan, O.; Narayan, K.S.; Goodwin, T.J.; Spaulding, G.F. Three-dimensional growth patterns of various human tumor cell lines in simulated microgravity of a NASA bioreactor. In Vitro Cell Dev. Biol. Anim. 1997, 33, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef]
- Caliari, S.R.; Burdick, J.A. A practical guide to hydrogels for cell culture. Nat. Methods 2016, 13, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Shoseyov, O.; Posen, Y.; Grynspan, F. Human collagen produced in plants: More than just another molecule. Bioengineered 2014, 5, 49–52. [Google Scholar] [CrossRef] [Green Version]
- Kraham, S.J. Environmental Impacts of Industrial Livestock Production. In International Farm Animal, Wildlife and Food Safety Law; Steier, G., Patel, K.K., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 3–40. [Google Scholar]
- Verbeke, W.A.J.; Viaene, J. Ethical Challenges for Livestock Production:Meeting Consumer Concerns about Meat Safety and AnimalWelfare. J. Agric. Environ. Ethics 2000, 12, 141–151. [Google Scholar] [CrossRef]
- Kuo, C.K.; Tuan, R.S. Mechanoactive tenogenic differentiation of human mesenchymal stem cells. Tissue Eng. Part A 2008, 14, 1615–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinman, H.K.; Martin, G.R. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 2005, 15, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.C.; Barker, T.H. Fibrin-based biomaterials: Modulation of macroscopic properties through rational design at the molecular level. Acta Biomater. 2014, 10, 1502–1514. [Google Scholar] [CrossRef] [Green Version]
- Del Bufalo, F.; Manzo, T.; Hoyos, V.; Yagyu, S.; Caruana, I.; Jacot, J.; Benavides, O.; Rosen, D.; Brenner, M.K. 3D modeling of human cancer: A PEG-fibrin hydrogel system to study the role of tumor microenvironment and recapitulate the in vivo effect of oncolytic adenovirus. Biomaterials 2016, 84, 76–85. [Google Scholar] [CrossRef]
- Paxton, J.Z.; Wudebwe, U.N.; Wang, A.; Woods, D.; Grover, L.M. Monitoring sinew contraction during formation of tissue-engineered fibrin-based ligament constructs. Tissue Eng. Part A 2012, 18, 1596–1607. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Malinen, M.M.; Lauren, P.; Lou, Y.R.; Kuisma, S.W.; Kanninen, L.; Lille, M.; Corlu, A.; GuGuen-Guillouzo, C.; Ikkala, O.; et al. Nanofibrillar cellulose hydrogel promotes three-dimensional liver cell culture. J. Control. Release 2012, 164, 291–298. [Google Scholar] [CrossRef]
- Oliveira, J.T.; Crawford, A.; Mundy, J.M.; Moreira, A.R.; Gomes, M.E.; Hatton, P.V.; Reis, R.L. A cartilage tissue engineering approach combining starch-polycaprolactone fibre mesh scaffolds with bovine articular chondrocytes. J. Mater. Sci. Mater. Med. 2007, 18, 295–302. [Google Scholar] [CrossRef]
- Van Nieuwenhove, I.; Salamon, A.; Adam, S.; Dubruel, P.; Van Vlierberghe, S.; Peters, K. Gelatin- and starch-based hydrogels. Part B: In vitro mesenchymal stem cell behavior on the hydrogels. Carbohydr. Polym. 2017, 161, 295–305. [Google Scholar] [CrossRef]
- Normand, V.; Lootens, D.L.; Amici, E.; Plucknett, K.P.; Aymard, P. New insight into agarose gel mechanical properties. Biomacromolecules 2000, 1, 730–738. [Google Scholar] [CrossRef]
- Sánchez-Salcedo, S.; Nieto, A.; Vallet-Regí, M. Hydroxyapatite/β-tricalcium phosphate/agarose macroporous scaffolds for bone tissue engineering. Chem. Eng. J. 2008, 137, 62–71. [Google Scholar] [CrossRef]
- Rabyk, M.; Hruby, M.; Vetrik, M.; Kucka, J.; Proks, V.; Parizek, M.; Konefal, R.; Krist, P.; Chvatil, D.; Bacakova, L.; et al. Modified glycogen as construction material for functional biomimetic microfibers. Carbohydr. Polym. 2016, 152, 271–279. [Google Scholar] [CrossRef]
- Gopinath, V.; Saravanan, S.; Al-Maleki, A.R.; Ramesh, M.; Vadivelu, J. A review of natural polysaccharides for drug delivery applications: Special focus on cellulose, starch and glycogen. Biomed Pharm. 2018, 107, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Rodrigues, J.; Tomas, H. Injectable and biodegradable hydrogels: Gelation, biodegradation and biomedical applications. Chem. Soc. Rev. 2012, 41, 2193–2221. [Google Scholar] [CrossRef]
- Zacchi, V.; Soranzo, C.; Cortivo, R.; Radice, M.; Brun, P.; Abatangelo, G. In vitro engineering of human skin-like tissue. J. Biomed. Mater. Res. 1998, 40, 187–194. [Google Scholar] [CrossRef]
- Webber, M.J.; Tongers, J.; Newcomb, C.J.; Marquardt, K.T.; Bauersachs, J.; Losordo, D.W.; Stupp, S.I. Supramolecular nanostructures that mimic VEGF as a strategy for ischemic tissue repair. Proc. Natl. Acad. Sci. USA 2011, 108, 13438–13443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kisiday, J.; Jin, M.; Kurz, B.; Hung, H.; Semino, C.; Zhang, S.; Grodzinsky, A.J. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: Implications for cartilage tissue repair. Proc. Natl. Acad. Sci. USA 2002, 99, 9996–10001. [Google Scholar] [CrossRef] [Green Version]
- Rowlands, A.S.; George, P.A.; Cooper-White, J.J. Directing osteogenic and myogenic differentiation of MSCs: Interplay of stiffness and adhesive ligand presentation. Am. J. Physiol. Cell Physiol. 2008, 295, C1037–C1044. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Takayama, S.; Park, J. Formation and manipulation of cell spheroids using a density adjusted PEG/DEX aqueous two phase system. Sci. Rep. 2015, 5, 11891. [Google Scholar] [CrossRef] [Green Version]
- Kojima, N.; Takeuchi, S.; Sakai, Y. Rapid aggregation of heterogeneous cells and multiple-sized microspheres in methylcellulose medium. Biomaterials 2012, 33, 4508–4514. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Tanner, K. Three-Dimensional Patterning of the ECM Microenvironment Using Magnetic Nanoparticle Self Assembly. Curr. Protoc. Cell Biol. 2016, 70, 25.3.1–25.3.14. [Google Scholar] [CrossRef]
- Urbanczyk, M.; Layland, S.L.; Schenke-Layland, K. The role of extracellular matrix in biomechanics and its impact on bioengineering of cells and 3D tissues. Matrix Biol. 2020, 85–86, 1–14. [Google Scholar] [CrossRef]
- Kacarevic, Z.P.; Rider, P.M.; Alkildani, S.; Retnasingh, S.; Smeets, R.; Jung, O.; Ivanisevic, Z.; Barbeck, M. An Introduction to 3D Bioprinting: Possibilities, Challenges and Future Aspects. Materials 2018, 11, 2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rider, P.; Kacarevic, Z.P.; Alkildani, S.; Retnasingh, S.; Barbeck, M. Bioprinting of tissue engineering scaffolds. J. Tissue Eng. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liao, K.; Li, C.; Lai, A.C.K.; Foo, J.J.; Chan, V. Progress in Integrative Biomaterial Systems to Approach Three-Dimensional Cell Mechanotransduction. Bioengineering 2017, 4, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burdett, E.; Kasper, F.K.; Mikos, A.G.; Ludwig, J.A. Engineering tumors: A tissue engineering perspective in cancer biology. Tissue Eng. Part B Rev. 2010, 16, 351–359. [Google Scholar] [CrossRef]
- Huebsch, N.; Gilbert, M.; Healy, K.E. Analysis of sterilization protocols for peptide-modified hydrogels. J. Biomed. Mater. Res. B Appl. Biomater. 2005, 74, 440–447. [Google Scholar] [CrossRef]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somaiah, C.; Kumar, A.; Mawrie, D.; Sharma, A.; Patil, S.D.; Bhattacharyya, J.; Swaminathan, R.; Jaganathan, B.G. Collagen Promotes Higher Adhesion, Survival and Proliferation of Mesenchymal Stem Cells. PLoS ONE 2015, 10, e0145068. [Google Scholar] [CrossRef] [Green Version]
- Weisel, J.W.; Litvinov, R.I. Mechanisms of fibrin polymerization and clinical implications. Blood 2013, 121, 1712–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litvinov, R.I.; Weisel, J.W. Fibrin mechanical properties and their structural origins. Matrix Biol. 2017, 60–61, 110–123. [Google Scholar] [CrossRef]
- Cheng, Y.; Luo, X.; Payne, G.; Rubloff, G. Biofabrication: Programmable assembly of polysaccharide hydrogels in microfluidics as biocompatible scaffolds. J. Mater. Chem. 2012, 22, 7659–7666. [Google Scholar] [CrossRef]
- Contessi, N.; Altomare, L.; Filipponi, A.; Farè, S. Thermo-responsive properties of methylcellulose hydrogels for cell sheet engineering. Mater. Lett. 2017, 207, 157–160. [Google Scholar] [CrossRef]
- Mukerjea, R.; Slocum, G.; Robyt, J.F. Determination of the maximum water solubility of eight native starches and the solubility of their acidic-methanol and -ethanol modified analogues. Carbohydr. Res. 2007, 342, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Ratnayake, W.S.; Jackson, D.S. Gelatinization and solubility of corn starch during heating in excess water: New insights. J. Agric. Food Chem. 2006, 54, 3712–3716. [Google Scholar] [CrossRef] [Green Version]
- Perrone, M.; Lopalco, A.; Lopedota, A.; Cutrignelli, A.; Laquintana, V.; Douglas, J.; Franco, M.; Liberati, E.; Russo, V.; Tongiani, S.; et al. Preactivated thiolated glycogen as mucoadhesive polymer for drug delivery. Eur. J. Pharm. Biopharm. 2017, 119, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Khetan, S.; Guvendiren, M.; Legant, W.R.; Cohen, D.M.; Chen, C.S.; Burdick, J.A. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat. Mater. 2013, 12, 458–465. [Google Scholar] [CrossRef] [Green Version]
- Burdick, J.A.; Prestwich, G.D. Hyaluronic acid hydrogels for biomedical applications. Adv. Mater. 2011, 23, H41–H56. [Google Scholar] [CrossRef]
- Guvendiren, M.; Burdick, J.A. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun. 2012, 3, 792. [Google Scholar] [CrossRef] [Green Version]
- Matson, J.B.; Stupp, S.I. Self-assembling peptide scaffolds for regenerative medicine. Chem. Commun. 2012, 48, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Pelham, R.J., Jr.; Wang, Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA 1997, 94, 13661–13665. [Google Scholar] [CrossRef] [Green Version]
- Damljanovic, V.; Lagerholm, B.C.; Jacobson, K. Bulk and micropatterned conjugation of extracellular matrix proteins to characterized polyacrylamide substrates for cell mechanotransduction assays. Biotechniques 2005, 39, 847–851. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.C.; Anseth, K.S. PEG hydrogels for the controlled release of biomolecules in regenerative medicine. Pharm. Res. 2009, 26, 631–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hehlgans, S.; Lange, I.; Eke, I.; Cordes, N. 3D cell cultures of human head and neck squamous cell carcinoma cells are radiosensitized by the focal adhesion kinase inhibitor TAE226. Radiother. Oncol. 2009, 92, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Cordes, N.; Meineke, V. Cell adhesion-mediated radioresistance (CAM-RR). Extracellular matrix-dependent improvement of cell survival in human tumor and normal cells in vitro. Strahlenther. Onkol. 2003, 179, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Olive, P.L.; Durand, R.E. Drug and radiation resistance in spheroids: Cell contact and kinetics. Cancer Metastasis Rev. 1994, 13, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, G.; Nicolas, V.; Matsusaki, M.; Akashi, M.; Couvreur, P.; Mura, S. Multicellular spheroid based on a triple co-culture: A novel 3D model to mimic pancreatic tumor complexity. Acta Biomater. 2018, 78, 296–307. [Google Scholar] [CrossRef]
- Weigelt, B.; Lo, A.T.; Park, C.C.; Gray, J.W.; Bissell, M.J. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res. Treat. 2010, 122, 35–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Z.; Liao, Q.; Hu, Y.; You, L.; Zhou, L.; Zhao, Y. A spheroid-based 3-D culture model for pancreatic cancer drug testing, using the acid phosphatase assay. Braz. J. Med. Biol. Res. 2013, 46, 634–642. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, A.Y.; Torisawa, Y.S.; Tung, Y.C.; Sud, S.; Taichman, R.S.; Pienta, K.J.; Takayama, S. Microfluidic system for formation of PC-3 prostate cancer co-culture spheroids. Biomaterials 2009, 30, 3020–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longati, P.; Jia, X.; Eimer, J.; Wagman, A.; Witt, M.R.; Rehnmark, S.; Verbeke, C.; Toftgard, R.; Lohr, M.; Heuchel, R.L. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer 2013, 13, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiss, D.L.; Windus, L.C.; Avery, V.M. Chemokine receptor expression on integrin-mediated stellate projections of prostate cancer cells in 3D culture. Cytokine 2013, 64, 122–130. [Google Scholar] [CrossRef]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef] [Green Version]
- Rebelo, S.P.; Pinto, C.; Martins, T.R.; Harrer, N.; Estrada, M.F.; Loza-Alvarez, P.; Cabecadas, J.; Alves, P.M.; Gualda, E.J.; Sommergruber, W.; et al. 3D-3-culture: A tool to unveil macrophage plasticity in the tumour microenvironment. Biomaterials 2018, 163, 185–197. [Google Scholar] [CrossRef]
- Cuccarese, M.F.; Dubach, J.M.; Pfirschke, C.; Engblom, C.; Garris, C.; Miller, M.A.; Pittet, M.J.; Weissleder, R. Heterogeneity of macrophage infiltration and therapeutic response in lung carcinoma revealed by 3D organ imaging. Nat. Commun. 2017, 8, 14293. [Google Scholar] [CrossRef] [Green Version]
- Zachari, M.A.; Chondrou, P.S.; Pouliliou, S.E.; Mitrakas, A.G.; Abatzoglou, I.; Zois, C.E.; Koukourakis, M.I. Evaluation of the alamarblue assay for adherent cell irradiation experiments. Dose Response 2014, 12, 246–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsolou, A.; Angelou, E.; Didaskalou, S.; Bikiaris, D.; Avgoustakis, K.; Agianian, B.; Koffa, M.D. Folate and Pegylated Aliphatic Polyester Nanoparticles for Targeted Anticancer Drug Delivery. Int. J. Nanomed. 2020, 15, 4899–4918. [Google Scholar] [CrossRef] [PubMed]
- Monico, D.A.; Calori, I.R.; Souza, C.; Espreafico, E.M.; Bi, H.; Tedesco, A.C. Melanoma spheroid-containing artificial dermis as an alternative approach to in vivo models. Exp. Cell Res. 2022, 417, 113207. [Google Scholar] [CrossRef] [PubMed]
- Gopal, S.; Kwon, S.J.; Ku, B.; Lee, D.W.; Kim, J.; Dordick, J.S. 3D tumor spheroid microarray for high-throughput, high-content natural killer cell-mediated cytotoxicity. Commun. Biol. 2021, 4, 893. [Google Scholar] [CrossRef]
- Tsolou, A.; Koparanis, D.; Lamprou, I.; Giatromanolaki, A.; Koukourakis, M.I. Increased glucose influx and glycogenesis in lung cancer cells surviving after irradiation. Int. J. Radiat. Biol. 2022, 99, 692–701. [Google Scholar] [CrossRef] [PubMed]
- Sreedhar, A.; Zhao, Y. Dysregulated metabolic enzymes and metabolic reprogramming in cancer cells. Biomed. Rep. 2018, 8, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivridis, E.; Giatromanolaki, A.; Koukourakis, M.I. “Stromatogenesis” and tumor progression. Int. J. Surg. Pathol. 2004, 12, 1–9. [Google Scholar] [CrossRef]
- Martinez-Outschoorn, U.E.; Pavlides, S.; Howell, A.; Pestell, R.G.; Tanowitz, H.B.; Sotgia, F.; Lisanti, M.P. Stromal-epithelial metabolic coupling in cancer: Integrating autophagy and metabolism in the tumor microenvironment. Int. J. Biochem. Cell Biol. 2011, 43, 1045–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koukourakis, M.I.; Kalamida, D.; Mitrakas, A.G.; Liousia, M.; Pouliliou, S.; Sivridis, E.; Giatromanolaki, A. Metabolic cooperation between co-cultured lung cancer cells and lung fibroblasts. Lab. Investig. 2017, 97, 1321–1331. [Google Scholar] [CrossRef] [Green Version]
- Liao, J.; Qian, F.; Tchabo, N.; Mhawech-Fauceglia, P.; Beck, A.; Qian, Z.; Wang, X.; Huss, W.J.; Lele, S.B.; Morrison, C.D.; et al. Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS ONE 2014, 9, e84941. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Lin, D.; Taniguchi, C.M. Hypoxia inducible factor (HIF) in the tumor microenvironment: Friend or foe? Sci. China Life Sci. 2017, 60, 1114–1124. [Google Scholar] [CrossRef] [PubMed]
- McKeown, S.R. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br. J. Radiol. 2014, 87, 20130676. [Google Scholar] [CrossRef] [Green Version]
- Han, S.J.; Kwon, S.; Kim, K.S. Challenges of applying multicellular tumor spheroids in preclinical phase. Cancer Cell Int. 2021, 21, 152. [Google Scholar] [CrossRef]
- Olofsson, K.; Carannante, V.; Takai, M.; Onfelt, B.; Wiklund, M. Single cell organization and cell cycle characterization of DNA stained multicellular tumor spheroids. Sci. Rep. 2021, 11, 17076. [Google Scholar] [CrossRef]
- Laurent, J.; Frongia, C.; Cazales, M.; Mondesert, O.; Ducommun, B.; Lobjois, V. Multicellular tumor spheroid models to explore cell cycle checkpoints in 3D. BMC Cancer 2013, 13, 73. [Google Scholar] [CrossRef] [Green Version]
- Sakaue-Sawano, A.; Kurokawa, H.; Morimura, T.; Hanyu, A.; Hama, H.; Osawa, H.; Kashiwagi, S.; Fukami, K.; Miyata, T.; Miyoshi, H.; et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008, 132, 487–498. [Google Scholar] [CrossRef] [Green Version]
- Beaumont, K.A.; Anfosso, A.; Ahmed, F.; Weninger, W.; Haass, N.K. Imaging-and Flow Cytometry-based Analysis of Cell Position and the Cell Cycle in 3D Melanoma Spheroids. J. Vis. Exp. 2015, 106, e53486. [Google Scholar]
- Murphy, R.J.; Browning, A.P.; Gunasingh, G.; Haass, N.K.; Simpson, M.J. Designing and interpreting 4D tumour spheroid experiments. Commun. Biol. 2022, 5, 91. [Google Scholar] [CrossRef] [PubMed]
- Browning, A.P.; Sharp, J.A.; Murphy, R.J.; Gunasingh, G.; Lawson, B.; Burrage, K.; Haass, N.K.; Simpson, M. Quantitative analysis of tumour spheroid structure. Elife 2021, 10, e73020. [Google Scholar] [CrossRef]
- Desmaison, A.; Frongia, C.; Grenier, K.; Ducommun, B.; Lobjois, V. Mechanical stress impairs mitosis progression in multi-cellular tumor spheroids. PLoS ONE 2013, 8, e80447. [Google Scholar] [CrossRef] [Green Version]
- Haass, N.K.; Beaumont, K.A.; Hill, D.S.; Anfosso, A.; Mrass, P.; Munoz, M.A.; Kinjyo, I.; Weninger, W. Real-time cell cycle imaging during melanoma growth, invasion, and drug response. Pigment. Cell Melanoma Res. 2014, 27, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, G.; Vinciguerra, D.; Balasso, A.; Nicolas, V.; Goudin, N.; Garfa-Traore, M.; Feher, A.; Dinnyes, A.; Nicolas, J.; Couvreur, P.; et al. Light sheet fluorescence microscopy versus confocal microscopy: In quest of a suitable tool to assess drug and nanomedicine penetration into multicellular tumor spheroids. Eur. J. Pharm. Biopharm. 2019, 142, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, C.; Frongia, C.; Jorand, R.; Fehrenbach, J.; Weiss, P.; Maandhui, A.; Gay, G.; Ducommun, B.; Lobjois, V. Live cell division dynamics monitoring in 3D large spheroid tumor models using light sheet microscopy. Cell Div. 2011, 6, 22. [Google Scholar] [CrossRef] [Green Version]
- Pampaloni, F.; Ansari, N.; Stelzer, E.H. High-resolution deep imaging of live cellular spheroids with light-sheet-based fluorescence microscopy. Cell Tissue Res. 2013, 352, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Eismann, B.; Krieger, T.G.; Beneke, J.; Bulkescher, R.; Adam, L.; Erfle, H.; Herrmann, C.; Eils, R.; Conrad, C. Automated 3D light-sheet screening with high spatiotemporal resolution reveals mitotic phenotypes. J. Cell Sci. 2020, 133, jcs245043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monjaret, F.; Fernandes, M.; Duchemin-Pelletier, E.; Argento, A.; Degot, S.; Young, J. Fully Automated One-Step Production of Functional 3D Tumor Spheroids for High-Content Screening. J. Lab. Autom. 2016, 21, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Mittler, F.; Obeid, P.; Rulina, A.V.; Haguet, V.; Gidrol, X.; Balakirev, M.Y. High-Content Monitoring of Drug Effects in a 3D Spheroid Model. Front. Oncol. 2017, 7, 293. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, D.P.; Parker, T.L.; Walker, D.A.; Alexander, C.; Ashford, M.B.; Gellert, P.R.; Garnett, M.C. Multiplexing spheroid volume, resazurin and acid phosphatase viability assays for high-throughput screening of tumour spheroids and stem cell neurospheres. PLoS ONE 2014, 9, e103817. [Google Scholar] [CrossRef] [Green Version]
- Kandilogiannakis, L.; Filidou, E.; Drygiannakis, I.; Tarapatzi, G.; Didaskalou, S.; Koffa, M.; Arvanitidis, K.; Bamias, G.; Valatas, V.; Paspaliaris, V.; et al. Development of a Human Intestinal Organoid Model for In Vitro Studies on Gut Inflammation and Fibrosis. Stem Cells Int. 2021, 2021, 9929461. [Google Scholar] [CrossRef]
- Smyrek, I.; Mathew, B.; Fischer, S.C.; Lissek, S.M.; Becker, S.; Stelzer, E.H.K. E-cadherin, actin, microtubules and FAK dominate different spheroid formation phases and important elements of tissue integrity. Biol. Open 2019, 8, bio037051. [Google Scholar] [CrossRef] [Green Version]
- Engelbrecht, C.J.; Stelzer, E.H. Resolution enhancement in a light-sheet-based microscope (SPIM). Opt. Lett. 2006, 31, 1477–1479. [Google Scholar] [CrossRef] [Green Version]
- Power, R.M.; Huisken, J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat. Methods 2017, 14, 360–373. [Google Scholar] [CrossRef]
- Reynaud, E.G.; Krzic, U.; Greger, K.; Stelzer, E.H. Light sheet-based fluorescence microscopy: More dimensions, more photons, and less photodamage. HFSP J. 2008, 2, 266–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huisken, J.; Swoger, J.; Del Bene, F.; Wittbrodt, J.; Stelzer, E.H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 2004, 305, 1007–1009. [Google Scholar] [CrossRef] [Green Version]
- Swoger, J.; Pampaloni, F.; Stelzer, E.H. Light-sheet-based fluorescence microscopy for three-dimensional imaging of biological samples. Cold Spring Harb. Protoc. 2014, 2014, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Huisken, J.; Stainier, D.Y. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt. Lett. 2007, 32, 2608–2610. [Google Scholar] [CrossRef]
- Dodt, H.U.; Leischner, U.; Schierloh, A.; Jahrling, N.; Mauch, C.P.; Deininger, K.; Deussing, J.M.; Eder, M.; Zieglgansberger, W.; Becker, K. Ultramicroscopy: Three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 2007, 4, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Tomer, R.; Khairy, K.; Amat, F.; Keller, P.J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 2012, 9, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.J.; Schmidt, A.D.; Wittbrodt, J.; Stelzer, E.H. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 2008, 322, 1065–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masson, A.; Escande, P.; Frongia, C.; Clouvel, G.; Ducommun, B.; Lorenzo, C. High-resolution in-depth imaging of optically cleared thick samples using an adaptive SPIM. Sci. Rep. 2015, 5, 16898. [Google Scholar] [CrossRef] [Green Version]
- Boutin, M.E.; Voss, T.C.; Titus, S.A.; Cruz-Gutierrez, K.; Michael, S.; Ferrer, M. A high-throughput imaging and nuclear segmentation analysis protocol for cleared 3D culture models. Sci. Rep. 2018, 8, 11135. [Google Scholar] [CrossRef] [Green Version]
- Desmaison, A.; Guillaume, L.; Triclin, S.; Weiss, P.; Ducommun, B.; Lobjois, V. Impact of physical confinement on nuclei geometry and cell division dynamics in 3D spheroids. Sci. Rep. 2018, 8, 8785. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, A.; Fischer, S.C.; Mattheyer, C.; Pampaloni, F.; Stelzer, E.H. Multiscale image analysis reveals structural heterogeneity of the cell microenvironment in homotypic spheroids. Sci. Rep. 2017, 7, 43693. [Google Scholar] [CrossRef]
- Beghin, A.; Grenci, G.; Sahni, G.; Guo, S.; Rajendiran, H.; Delaire, T.; Mohamad Raffi, S.B.; Blanc, D.; de Mets, R.; Ong, H.T.; et al. Automated high-speed 3D imaging of organoid cultures with multi-scale phenotypic quantification. Nat. Methods 2022, 19, 881–892. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [Green Version]
- Arganda-Carreras, I.; Kaynig, V.; Rueden, C.; Eliceiri, K.W.; Schindelin, J.; Cardona, A.; Sebastian Seung, H. Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 2017, 33, 2424–2426. [Google Scholar] [CrossRef]
- Tischer, C.; Pepperkok, R. CATS: Fiji Plugin for Context Aware Trainable Segmentation for Big Image Data (v0.5.01). Zenodo. 2017. Available online: https://zenodo.org/record/2600293#.ZC_q6XZBxPY (accessed on 30 January 2023).
- Arzt, M.; Deschamps, J.; Schmied, C.; Pietzsch, T.; Schmidt, D.; Tomancak, P.; Haase, R.; Jug, F. LABKIT: Labeling and Segmentation Toolkit for Big Image Data. Front. Comput. Sci. 2022, 4, 777728. [Google Scholar] [CrossRef]
- Pietzsch, T.; Saalfeld, S.; Preibisch, S.; Tomancak, P. BigDataViewer: Visualization and processing for large image data sets. Nat. Methods 2015, 12, 481–483. [Google Scholar] [CrossRef]
- Cao, J.; Guan, G.; Ho, V.W.S.; Wong, M.K.; Chan, L.Y.; Tang, C.; Zhao, Z.; Yan, H. Establishment of a morphological atlas of the Caenorhabditis elegans embryo using deep-learning-based 4D segmentation. Nat. Commun. 2020, 11, 6254. [Google Scholar] [CrossRef] [PubMed]
- Richards, R.; Mason, D.; Lévy, R.; Bearon, R.; Sée, V. 4D imaging and analysis of multicellular tumour spheroid cell migration and invasion. bioRxiv 2018, 443648. [Google Scholar] [CrossRef] [Green Version]
- Nasser, L.; Boudier, T. A novel generic dictionary-based denoising method for improving noisy and densely packed nuclei segmentation in 3D time-lapse fluorescence microscopy images. Sci. Rep. 2019, 9, 5654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. Ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef]
- Alladin, A.; Chaible, L.; Garcia Del Valle, L.; Sabine, R.; Loeschinger, M.; Wachsmuth, M.; Heriche, J.K.; Tischer, C.; Jechlinger, M. Tracking cells in epithelial acini by light sheet microscopy reveals proximity effects in breast cancer initiation. Elife 2020, 9, e54066. [Google Scholar] [CrossRef]
- Stringer, C.; Wang, T.; Michaelos, M.; Pachitariu, M. Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 2021, 18, 100–106. [Google Scholar] [CrossRef]
- Weigert, M.; Schmidt, U.; Haase, R.; Sugawara, K.; Myers, G. Star-Convex Polyhedra for 3D Object Detection and Segmentation in Microscopy. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV 2020), Waikoloa, HI, USA, 3–8 January 2022. [Google Scholar]
- von Chamier, L.; Laine, R.F.; Jukkala, J.; Spahn, C.; Krentzel, D.; Nehme, E.; Lerche, M.; Hernandez-Perez, S.; Mattila, P.K.; Karinou, E.; et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat. Commun. 2021, 12, 2276. [Google Scholar] [CrossRef]
- Redmond, J.; McCarthy, H.; Buchanan, P.; Levingstone, T.J.; Dunne, N.J. Advances in biofabrication techniques for collagen-based 3D in vitro culture models for breast cancer research. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 122, 111944. [Google Scholar] [CrossRef]
- Serban, M.A.; Prestwich, G.D. Modular extracellular matrices: Solutions for the puzzle. Methods 2008, 45, 93–98. [Google Scholar] [CrossRef] [Green Version]
- Jeon, O.; Marks, R.; Wolfson, D.; Alsberg, E. Dual-crosslinked hydrogel microwell system for formation and culture of multicellular human adipose tissue-derived stem cell spheroids. J. Mater. Chem. B 2016, 4, 3526–3533. [Google Scholar] [CrossRef]
- Tseng, T.C.; Wong, C.W.; Hsieh, F.Y.; Hsu, S.H. Biomaterial Substrate-Mediated Multicellular Spheroid Formation and Their Applications in Tissue Engineering. Biotechnol. J. 2017, 12, 1700064. [Google Scholar] [CrossRef]
- Chen, X.; Thibeault, S.L. Cell-cell interaction between vocal fold fibroblasts and bone marrow mesenchymal stromal cells in three-dimensional hyaluronan hydrogel. J. Tissue Eng. Regen. Med. 2016, 10, 437–446. [Google Scholar] [CrossRef] [Green Version]
- Engel, B.J.; Constantinou, P.E.; Sablatura, L.K.; Doty, N.J.; Carson, D.D.; Farach-Carson, M.C.; Harrington, D.A.; Zarembinski, T.I. Multilayered, Hyaluronic Acid-Based Hydrogel Formulations Suitable for Automated 3D High Throughput Drug Screening of Cancer-Stromal Cell Cocultures. Adv. Healthc. Mater. 2015, 4, 1664–1674. [Google Scholar] [CrossRef]
- Mannino, R.G.; Santiago-Miranda, A.N.; Pradhan, P.; Qiu, Y.; Mejias, J.C.; Neelapu, S.S.; Roy, K.; Lam, W.A. 3D microvascular model recapitulates the diffuse large B-cell lymphoma tumor microenvironment in vitro. Lab Chip 2017, 17, 407–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, K.; Pollock, J.F.; Schaffer, D.V.; Healy, K.E. Designing synthetic materials to control stem cell phenotype. Curr. Opin. Chem. Biol. 2007, 11, 381–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Milleret, V.; Thompson-Steckel, G.; Huang, N.P.; Voros, J.; Simona, B.R.; Ehrbar, M. Soft Hydrogels Featuring In-Depth Surface Density Gradients for the Simple Establishment of 3D Tissue Models for Screening Applications. SLAS Discov. 2017, 22, 635–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nallanthighal, S.; Heiserman, J.P.; Cheon, D.J. The Role of the Extracellular Matrix in Cancer Stemness. Front. Cell Dev. Biol. 2019, 7, 86. [Google Scholar] [CrossRef] [Green Version]
- Saraswathibhatla, A.; Indana, D.; Chaudhuri, O. Cell-extracellular matrix mechanotransduction in 3D. Nat. Rev. Mol. Cell Biol. 2023, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Dorrigiv, D.; Goyette, P.A.; St-Georges-Robillard, A.; Mes-Masson, A.M.; Gervais, T. Pixelated Microfluidics for Drug Screening on Tumour Spheroids and Ex Vivo Microdissected Tumour Explants. Cancers 2023, 15, 1060. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Park, S.; Kim, J.E.; Park, B.; Hong, Y.; Lim, J.W.; Jeong, S.; Son, H.; Kim, H.B.; Seonwoo, H.; et al. Development of a Scaffold-on-a-Chip Platform to Evaluate Cell Infiltration and Osteogenesis on the 3D-Printed Scaffold for Bone Regeneration. ACS Biomater. Sci. Eng. 2023, 9, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Limongi, T.; Guzzi, F.; Parrotta, E.; Candeloro, P.; Scalise, S.; Lucchino, V.; Gentile, F.; Tirinato, L.; Coluccio, M.L.; Torre, B.; et al. Microfluidics for 3D Cell and Tissue Cultures: Microfabricative and Ethical Aspects Updates. Cells 2022, 11, 1699. [Google Scholar] [CrossRef]
- Watson, S.A.; Javanmardi, Y.; Zanieri, L.; Shahreza, S.; Ragazzini, R.; Bonfanti, P.; Moeendarbary, E. Integrated role of human thymic stromal cells in hematopoietic stem cell extravasation. Bioeng. Transl. Med. 2023, 8, e10454. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Halldorsson, S.; Lucumi, E.; Gomez-Sjoberg, R.; Fleming, R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shemesh, J.; Jalilian, I.; Shi, A.; Heng Yeoh, G.; Knothe Tate, M.L.; Ebrahimi Warkiani, M. Flow-induced stress on adherent cells in microfluidic devices. Lab Chip 2015, 15, 4114–4127. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Hass, R.; von der Ohe, J.; Ungefroren, H. Impact of the Tumor Microenvironment on Tumor Heterogeneity and Consequences for Cancer Cell Plasticity and Stemness. Cancers 2020, 12, 3716. [Google Scholar] [CrossRef]
- Joshi, A.; Choudhury, S.; Gugulothu, S.B.; Visweswariah, S.S.; Chatterjee, K. Strategies to Promote Vascularization in 3D Printed Tissue Scaffolds: Trends and Challenges. Biomacromolecules 2022, 23, 2730–2751. [Google Scholar] [CrossRef] [PubMed]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, S.; McOlash, L.; Palen, K.; Johnson, B.; Duris, C.; Yang, Q.; Dwinell, M.B.; Hunt, B.; Evans, D.B.; Gershan, J.; et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer 2018, 18, 335. [Google Scholar] [CrossRef]
- Aleman, J.; Skardal, A. A multi-site metastasis-on-a-chip microphysiological system for assessing metastatic preference of cancer cells. Biotechnol. Bioeng. 2019, 116, 936–944. [Google Scholar] [CrossRef]
- Caballero, D.; Kaushik, S.; Correlo, V.M.; Oliveira, J.M.; Reis, R.L.; Kundu, S.C. Organ-on-chip models of cancer metastasis for future personalized medicine: From chip to the patient. Biomaterials 2017, 149, 98–115. [Google Scholar] [CrossRef] [PubMed]





| 3D System | Advantages | Disadvantages |
|---|---|---|
| Liquid overlay cultures [29] |
|
|
| Hanging drop assays [30] |
|
|
| Spinner flasks [28,34] |
|
|
| Rotational culture systems [36] |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitrakas, A.G.; Tsolou, A.; Didaskalou, S.; Karkaletsou, L.; Efstathiou, C.; Eftalitsidis, E.; Marmanis, K.; Koffa, M. Applications and Advances of Multicellular Tumor Spheroids: Challenges in Their Development and Analysis. Int. J. Mol. Sci. 2023, 24, 6949. https://doi.org/10.3390/ijms24086949
Mitrakas AG, Tsolou A, Didaskalou S, Karkaletsou L, Efstathiou C, Eftalitsidis E, Marmanis K, Koffa M. Applications and Advances of Multicellular Tumor Spheroids: Challenges in Their Development and Analysis. International Journal of Molecular Sciences. 2023; 24(8):6949. https://doi.org/10.3390/ijms24086949
Chicago/Turabian StyleMitrakas, Achilleas G., Avgi Tsolou, Stylianos Didaskalou, Lito Karkaletsou, Christos Efstathiou, Evgenios Eftalitsidis, Konstantinos Marmanis, and Maria Koffa. 2023. "Applications and Advances of Multicellular Tumor Spheroids: Challenges in Their Development and Analysis" International Journal of Molecular Sciences 24, no. 8: 6949. https://doi.org/10.3390/ijms24086949