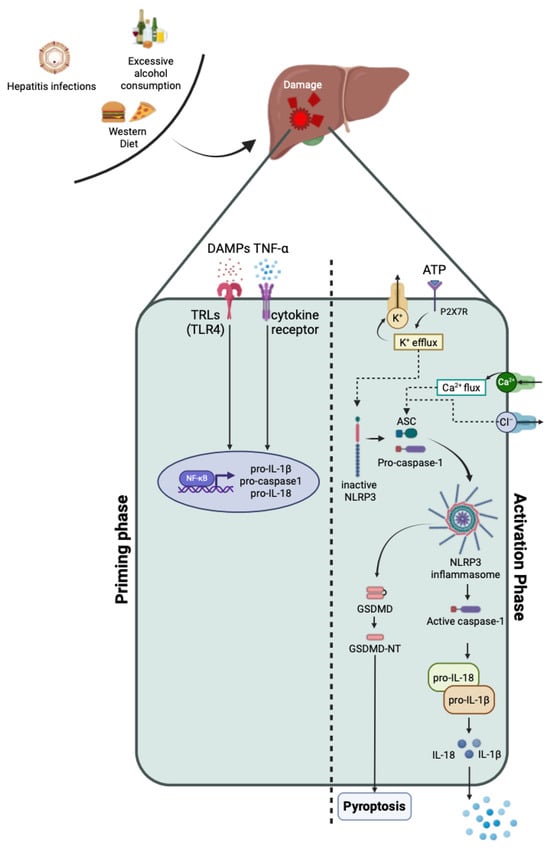
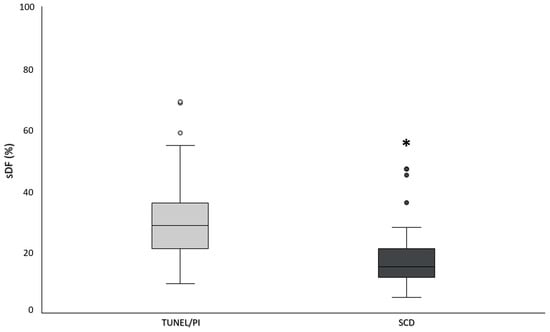
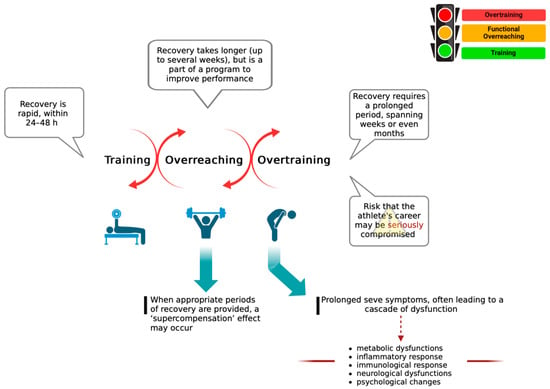
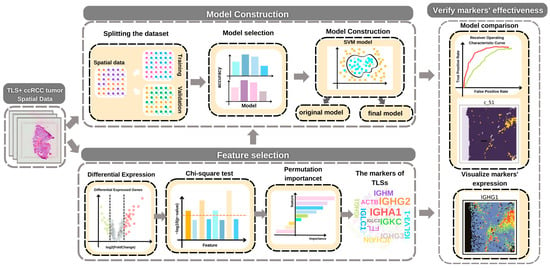
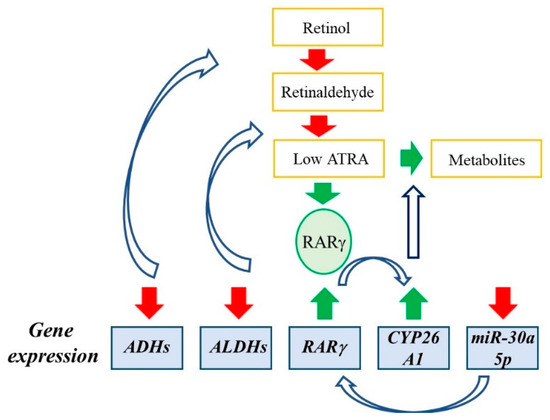
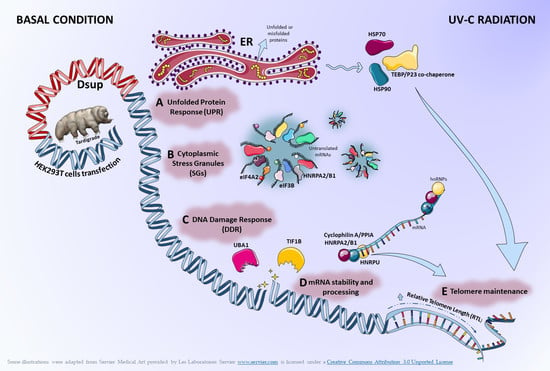
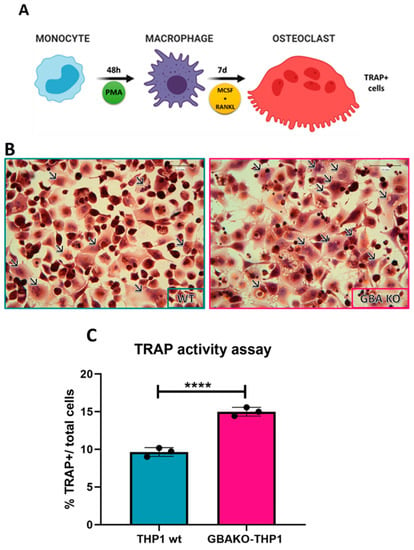
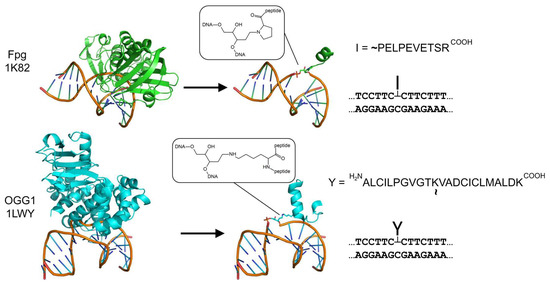
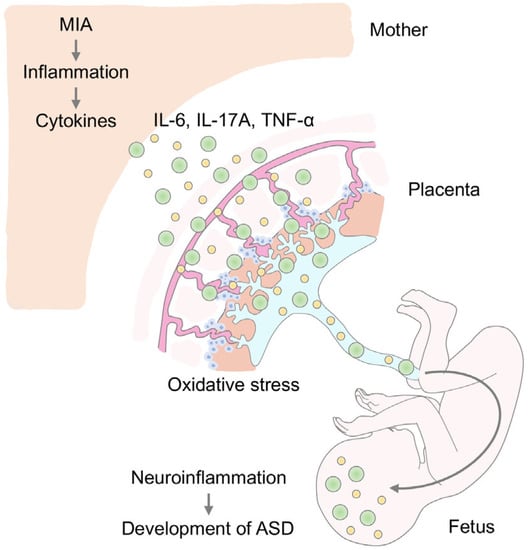
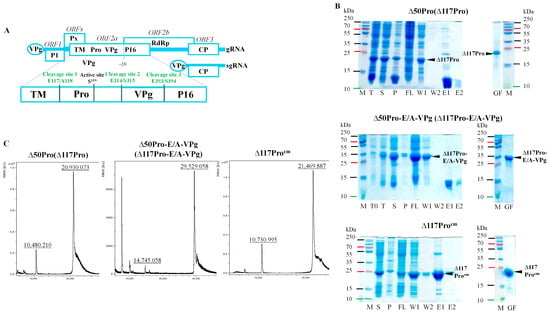
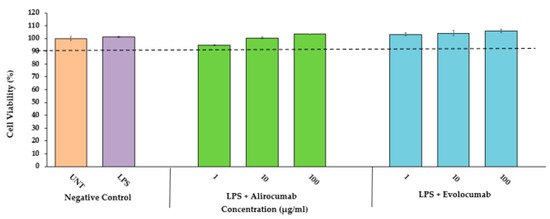
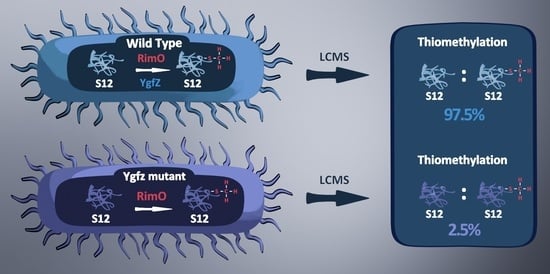
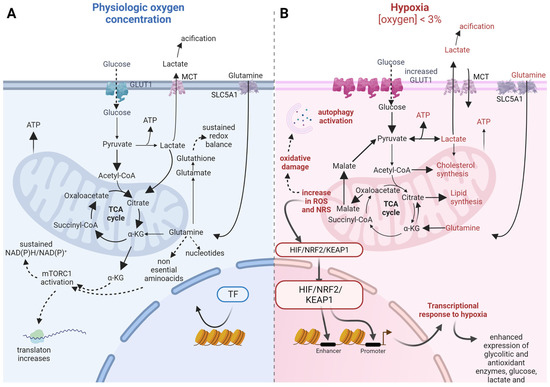
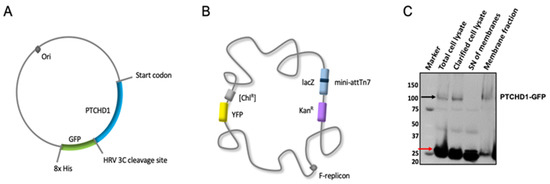
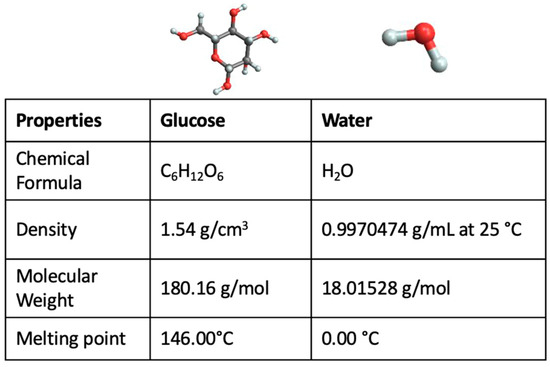
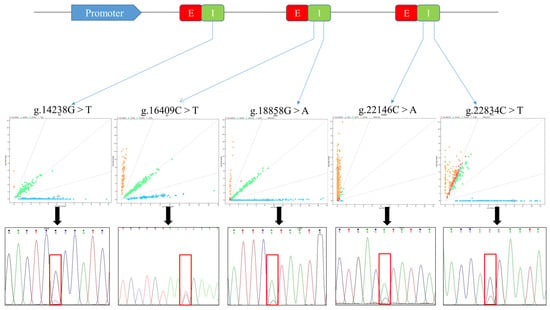
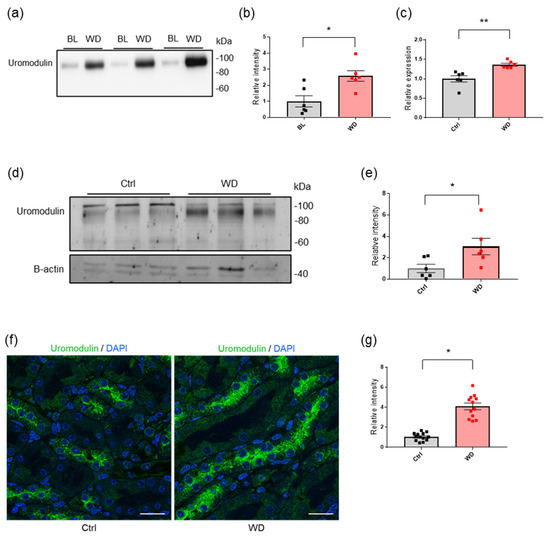
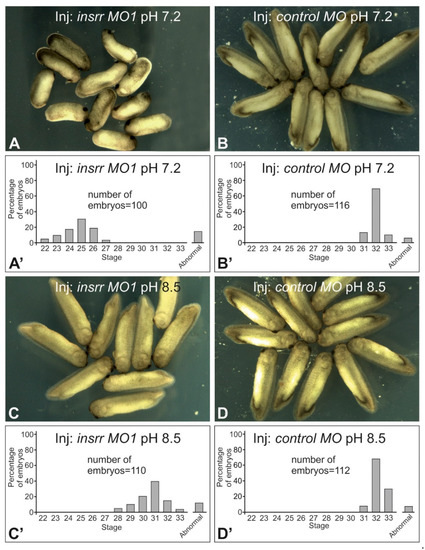
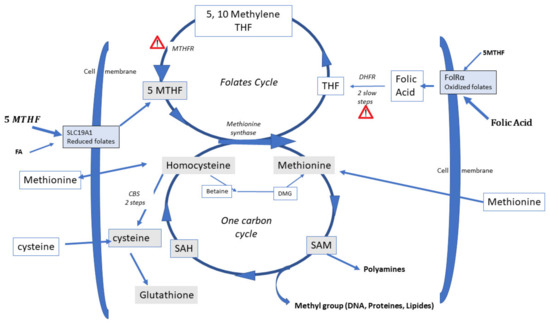
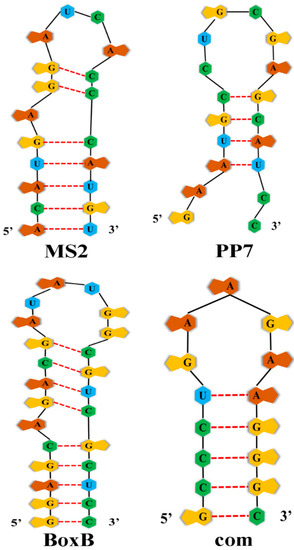
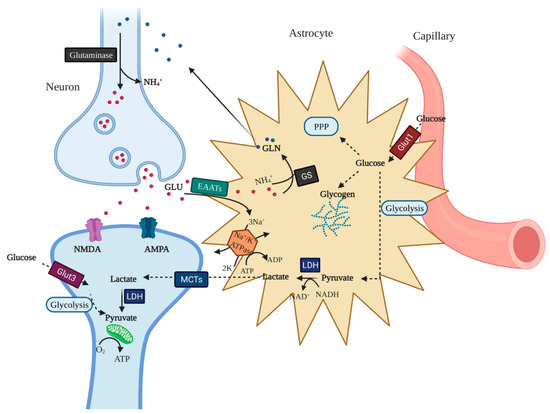
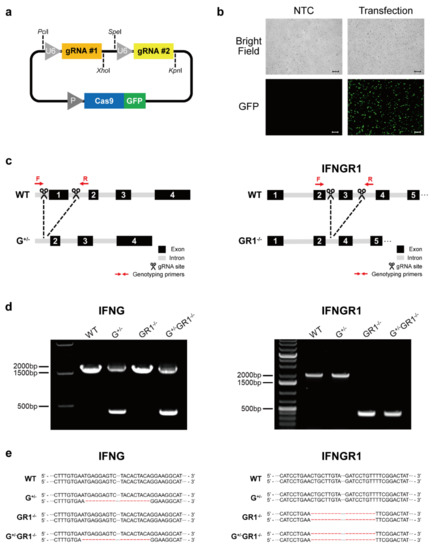
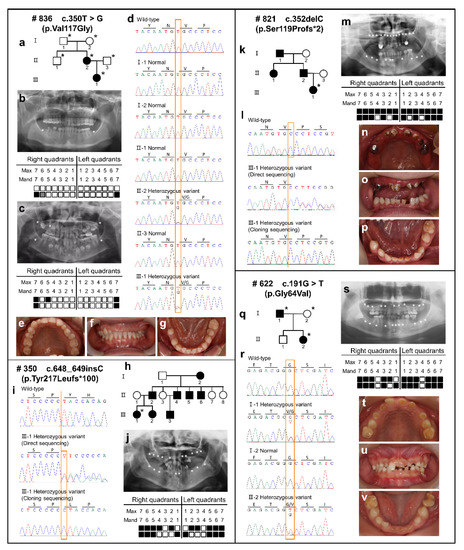
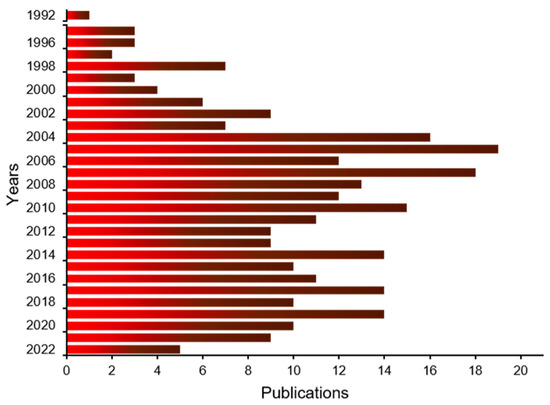
Feature Papers in “Molecular Biology”
A topical collection in International Journal of Molecular Sciences (ISSN 1422-0067). This collection belongs to the section "Molecular Biology".
Viewed by 495181Editor
Interests: nerve injury and neuropathic pain; pain and aging; central adaptations to chronic pain; multiple sclerosis; neuroinflammation; neuro-immunologic communication; redox signaling; nitric oxide; endocannabinoids and other lipid signaling molecules; progranulin; autophagy
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear colleagues,
This Topical Collection entitled “Feature Papers in “Molecular Biology”” aims to collect high-quality research articles, communications, and review articles in cutting-edge fields of molecular biology. Since the aim of this Topical Collection is to illustrate, through selected works, frontier research in molecular biology, we encourage Editorial Board Members of the Molecular Biology Section of the International Journal of Molecular Sciences to contribute feature papers reflecting the latest progress in their research field, or to invite relevant experts and colleagues to do so.
Topics include, but are not limited to,
- Biological activities at the molecular level;
- Biological processes of cell functions and maintenance;
- Molecular processes of cell division, senescence and cell death;
- Biomolecules interactions and cell-to-cell communication;
- DNA, and RNA biosynthesis, metabolism, interactions and functions;
- Protein biosynthesis, degradation and functions;
- Regulation of molecular interactions in a cell;
- Molecular processes of cell and organelle dynamics;
- Gene functions, genetics, and genomics;
- Signaling networks and system biology;
- Advanced techniques of molecular biology;
- Structural biology;
- Developmental biology;
- Chemical biology;
- Computational biology;
- Molecular methodologies, imaging techniques, and bioinformatics.
Prof. Dr. Irmgard Tegeder
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. International Journal of Molecular Sciences is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. There is an Article Processing Charge (APC) for publication in this open access journal. For details about the APC please see here. Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- molecular biology
- cell biology
- signal transduction and regulation
- cell growth and differentiation
- apoptosis
- necroptosis, ferroptosis, autophagy
- cell cycle
- macromolecules and complexes
- gene expression, functions and therapy
- mutation
- DNA structure, damage and repair
- chromatin structure and function
- nuclear organization
- polymerase
- RNA transcription
- RNA structure and regulation
- RNA splicing, and polyadenylation
- protein translation
- post-translational modifications
- amino acid
- proteins
- protein folding, chaperones, protein degradation and quality control
- enzyme regulation
- nucleic acid-protein interactions
- cell-cell interaction
- molecular clone
- sequencing analysis
- epigenetics
- proteomics
- bioinformatics
- imaging techniques
- kinesins
- metabolome
- transport mechanisms
- exosome, biological membranes
- homeostasis
Planned Papers
The below list represents only planned manuscripts. Some of these manuscripts have not been received by the Editorial Office yet. Papers submitted to MDPI journals are subject to peer-review.
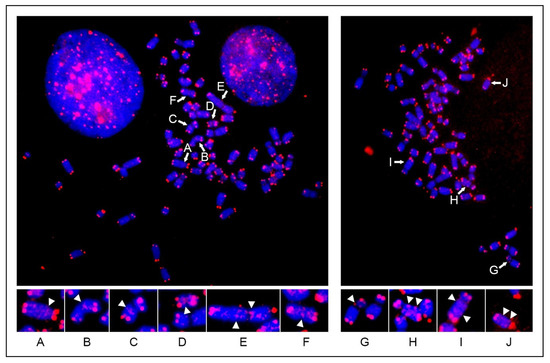
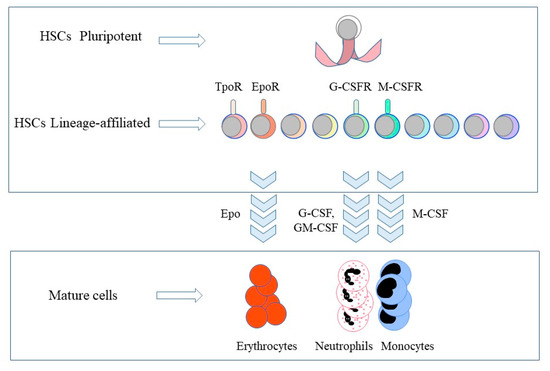
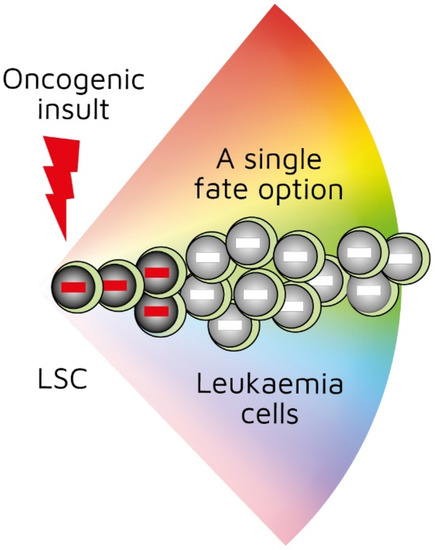
Title: Are cancer stems cells restricted to just a single cell lineage?
Authors: Geoffrey Brown1 and Isidro Sánchez-García2,3
1 Institute of Clinical Sciences, University of Birmingham, UK; 2Experimental Therapeutics and Translational Oncology Program, Instituto de Biología Molecular y Celular del Cáncer, CSIC/Universidad de Salamanca; 3Institute of Biomedical Research of Salamanca (IBSAL), Salamanca, Spain
Abstract: The cancer stem cell theory, which states that most if not all cancers arise from a stem cell, revolutionised the way we view cancer. Just as for normal cell development, cancer cells are a hierarchy of cells. The elegant studies of twins that both develop childhood acute lymphoblastic leukaemia revealed that at least two genomic insults are required for cancer. They, or more, ‘hits’ do not appear to confer a growth advantage to cancer cells, nor do cancer cells appear to be better equipped to survive than normal cells. Cancer cells in general and created by a specific genomic insult generally belong to just one cell lineage. In keeping with this and for example, transgenic mice in which the LMO2 (associated to human T-acute lymphoblastic leukaemia) and BCR-ABLp210 (associated to human chronic myeloid leukaemia) oncogenes were active solely within the haematopoietic stem cell compartment developed T lymphocyte and neutrophil lineage-restricted leukaemias respectively, typifying the human diseases. This ‘hard-wiring’ of lineage affiliation, either throughout leukaemia stem cell development or at a particular stage, is different to the behaviour of the normal haematopoietic stem cell. Whilst these cells can commit directly to a developmental pathway, they remain versatile and can still choose to develop towards an end cell type that is different from their initial commitment. We think that cancer stem cells do not have this versatility and that this is an essential difference between normal and cancer stem cells. Here we review the findings that support this notion.