Ca2+-Triggered Coelenterazine-Binding Protein Renilla: Expected and Unexpected Features
Abstract
:1. Introduction
2. Results
2.1. Theoretical Evaluations
2.2. Experimental Results
2.2.1. Furimazine-apoCBP Complex Preparation and Properties
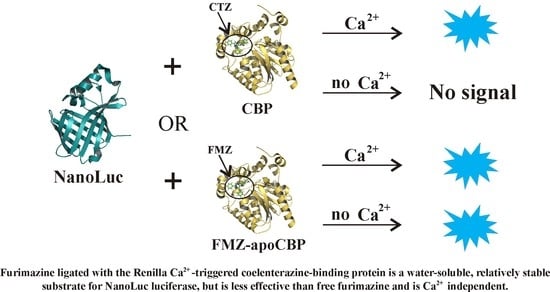
2.2.2. Bioluminescence NanoLuc Assay
3. Discussion
4. Materials and Methods
4.1. NanoLuc Expression and Purification
4.2. Furimazine-apoCBP Complex Production
4.3. Bioluminescence Assay Was Performed in Two Ways
4.4. Spectral Measurements
4.5. Theoretical Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Charbonneau, H.; Cormier, M.J. Ca2+-induced bioluminescence in Renilla reniformis. Purification and characterization of calcium triggered luciferin-binding protein. J. Biol. Chem. 1979, 254, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Stepanyuk, G.A.; Liu, Z.J.; Vysotski, E.S.; Lee, J.; Rose, J.P.; Wang, B.C. Structure based mechanism of the Ca2+-induced release of coelenterazine from the Renilla binding protein. Proteins 2009, 74, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Markova, S.V.; Vysotski, E.S. Coelenterazine-dependent luciferases. Biochemistry 2015, 80, 714–732. [Google Scholar] [CrossRef] [PubMed]
- Krasitskaya, V.V.; Bashmakova, E.E.; Frank, L.A. Coelenterazine-dependent luciferases as a powerful analytical tool for research and biomedical applications. Int. J. Mol. Sci. 2020, 21, 7465. [Google Scholar] [CrossRef]
- Coutant, E.P.; Goyard, S.; Hervin, V.; Gagnot, G.; Baatallah, R.; Jacob, Y.; Rose, T.; Janin, Y. Gram-scale synthesis of luciferins derived from coelenterazine and original insight into their bioluminescence properties. Org. Biomol. Chem. 2019, 17, 3709–3713. [Google Scholar] [CrossRef]
- Coutant, E.P.; Gagnot, G.; Hervin, V.; Baatallah, R.; Goyard, S.; Jacob, Y.; Rose, T.; Janin, Y.L. Bioluminescence profiling of NanoKAZ/NanoLuc luciferase using a chemical library of coelenterazine analogues. Chem. Eur. J. 2020, 26, 948–958. [Google Scholar] [CrossRef]
- Kudryavtsev, A.N.; Burakova, L.P.; Frank, L.A. Bioluminescent detection of tick-borne encephalitis virus in native ticks. Anal. Methods 2017, 9, 2252–2255. [Google Scholar] [CrossRef]
- Krasitskaya, V.V.; Burakova, L.P.; Pyshnaya, I.A.; Frank, L.A. Bioluminescent reporters for identification of gene allelic variants. Russ. J. Bioorgan. Chem. 2012, 38, 298–305. [Google Scholar] [CrossRef]
- Borisova, V.V.; Frank, L.A.; Markova, S.V.; Burakova, L.P.; Vysotski, E.S. Recombinant Metridia luciferase isoforms: Expression, refolding and applicability for in vitro assay. Photochem. Photobiol. Sci. 2008, 7, 1025–1031. [Google Scholar] [CrossRef]
- Titushin, M.S.; Markova, S.V.; Frank, L.A.; Malikova, N.P.; Stepanyuk, G.A.; Lee, J.; Vysotski, E.S. Coelenterazine-binding protein of Renilla muelleri: cDNA cloning, overexpression, and characterization as a substrate of luciferase. Photochem. Photobiol. Sci. 2008, 7, 189–196. [Google Scholar] [CrossRef]
- Stepanyuk, G.A.; Unch, J.; Malikova, N.P.; Markova, S.V.; Lee, J.; Vysotski, E.S. Coelenterazine-v ligated to Ca2+-triggered coelenterazine-binding protein is a stable and efficient substrate of the red-shifted mutant of Renilla muelleri luciferase. Anal. Bioanal. Chem. 2010, 398, 1809–1817. [Google Scholar] [CrossRef]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef]
- Peach, M.J.G.; Benfield, P.; Helgaker, T.; Tozer, D.J. Excitation energies in density functional theory: An evaluation and a diagnostic test. J. Chem. Phys. 2008, 128, 044118. [Google Scholar] [CrossRef]
- Tomasi, J.; Mennucci, B.; Cammi, R. Quantum mechanical continuum solvation models. Chem. Rev. 2005, 105, 2999–3094. [Google Scholar] [CrossRef]
- Tomilin, F.N.; Rogova, A.V.; Burakova, L.P.; Tchaikovskaya, O.N.; Avramov, P.V.; Fedorov, D.G.; Vysotski, E.S. Unusual shift in the visible absorption spectrum of an active ctenophore photoprotein elucidated by time-dependent density functional theory. Photochem. Photobiol. Sci. 2021, 20, 559–570. [Google Scholar] [CrossRef]
- Marques, M.A.; Gross, E.K. Time-dependent density functional theory. Annu. Rev. 2004, 55, 427–455. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.W.; Baldridge, K.K.; Boatz, J.A.; Elbert, S.T.; Gordon, M.S.; Jensen, J.H.; Koseki, S.; Matsunaga, N.; Nguyen, K.A.; Su, S.J.; et al. General atomic and molecular electronic structure system. J. Comput. Chem. 1993, 14, 1347–1363. [Google Scholar] [CrossRef]
- Fedorov, D.G. The fragment molecular orbital method: Theoretical development, implementation in GAMESS, and applications. Comput. Mol. Sci. 2017, 7, e1322. [Google Scholar] [CrossRef]
- Fedorov, D.G.; Kitaura, K. The importance of three-body terms in the fragment molecular orbital method. J. Chem. Phys. 2004, 120, 6832–6840. [Google Scholar] [CrossRef]
- Gaus, M.; Goez, A.; Elstner, M. Parametrization and Benchmark of DFTB3 for Organic Molecules. J. Chem. Theory Comput. 2012, 9, 338–354. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Fedorov, D.G. The fragment molecular orbital method combined with density-functional tight-binding and the polarizable continuum model. Phys. Chem. Chem. Phys. 2016, 18, 22047–22061. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, D.G. Partition analysis for density-functional tight-binding. J. Chem. Theory Comput. 2020, 124, 10346–10358. [Google Scholar] [CrossRef] [PubMed]
- Sladek, V.; Fedorov, D.G. The importance of charge transfer and solvent screening in the interactions of backbones and functional groups in amino acid residues and nucleotides. Int. J. Mol. Sci. 2022, 23, 13514. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, D.G.; Li, H.; Mironov, V.; Alexeev, Y. Computational Methods for Biochemical Simulations Implemented in GAMESS. In Quantum Mechanics in Drug Discovery, 2nd ed.; Heifetz, A., Ed.; Evotec: Abingdon, UK, 2020; Volume 2114, pp. 123–142. [Google Scholar]




| Conditions | Freshly Prepared, % | In 45 Days, % |
|---|---|---|
| 20 mM TrisHCl pH 7.0, 5 mM EDTA, 8 °C | 100 | 68.4 ± 0.6 |
| Frozen and thawed | 85.8 ± 0.2 | 85.6 ± 0.5 |
| Freeze-dried—dissolved | 68.2 ± 1.5 | 62.5 ± 0.5 |
| Freeze-dried (plus 0.1% BSA) | 96.4 ± 0.3 | 92.7 ± 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudryavtsev, A.N.; Krasitskaya, V.V.; Efremov, M.K.; Zangeeva, S.V.; Rogova, A.V.; Tomilin, F.N.; Frank, L.A. Ca2+-Triggered Coelenterazine-Binding Protein Renilla: Expected and Unexpected Features. Int. J. Mol. Sci. 2023, 24, 2144. https://doi.org/10.3390/ijms24032144
Kudryavtsev AN, Krasitskaya VV, Efremov MK, Zangeeva SV, Rogova AV, Tomilin FN, Frank LA. Ca2+-Triggered Coelenterazine-Binding Protein Renilla: Expected and Unexpected Features. International Journal of Molecular Sciences. 2023; 24(3):2144. https://doi.org/10.3390/ijms24032144
Chicago/Turabian StyleKudryavtsev, Alexander N., Vasilisa V. Krasitskaya, Maxim K. Efremov, Sayana V. Zangeeva, Anastasia V. Rogova, Felix N. Tomilin, and Ludmila A. Frank. 2023. "Ca2+-Triggered Coelenterazine-Binding Protein Renilla: Expected and Unexpected Features" International Journal of Molecular Sciences 24, no. 3: 2144. https://doi.org/10.3390/ijms24032144