Differentially Expressed Genes in Response to a Squalene-Supplemented Diet Are Accurate Discriminants of Porcine Non-Alcoholic Steatohepatitis
Abstract
:1. Introduction
2. Results
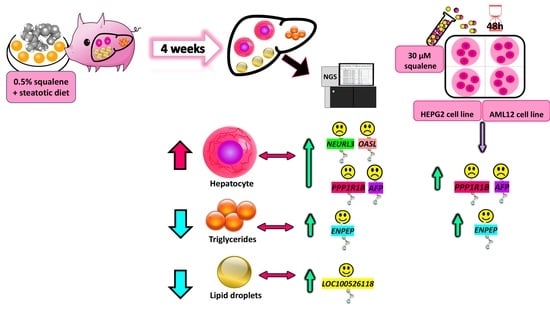
2.1. Hepatic Histological Analyses
2.2. Hepatic Gene Expression
2.3. Association among Gene Expression Changes and Pathological Features of NASH
2.4. Squalene Accumulates in the Liver and Is Responsible for the Changes in Gene Expressions
2.5. Squalene Is Responsible for the Changes in Genes Expression in Human Hepatoma G2 (HEPG2) and Murine Alpha Mouse Liver 12 (AML12) Cell Lines
3. Discussion
4. Materials and Methods
4.1. Animal Models and Experimental Design
4.2. Liver Histological Analyses
4.3. Quantification of Hepatic Lipids and Squalene
4.4. RNA Extraction
4.5. RNAseq and Data Analyses
4.6. Reverse Transcriptase-Quantitative PCR (RT-qPCR)
4.7. AML12 Cell Culture
4.8. HEPG2 Cell Culture
4.9. Quality Control and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Di Ciaula, A.; Passarella, S.; Shanmugam, H.; Noviello, M.; Bonfrate, L.; Wang, D.Q.-H.; Portincasa, P. Nonalcoholic fatty liver disease (NAFLD). Mitochondria as players and targets of therapies? Int. J. Mol. Sci. 2021, 22, 5375. [Google Scholar] [CrossRef]
- Bellentani, S. The epidemiology of non-alcoholic fatty liver disease. Liver Int. 2017, 37, 81–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, C.D.; Stengel, J.; Asike, M.I.; Torres, D.M.; Shaw, J.; Contreras, M.; Landt, C.L.; Harrison, S.A. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: A prospective study. Gastroenterology 2011, 140, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Dufour, J.-F.; Scherer, R.; Balp, M.-M.; McKenna, S.J.; Janssens, N.; Lopez, P.; Pedrosa, M. The global epidemiology of nonalcoholic steatohepatitis (NASH) and associated risk factors–A targeted literature review. Endocr. Metab. Sci. 2021, 3, 100089. [Google Scholar] [CrossRef]
- Bedossa, P.; Consortium, T.F.P. Utility and appropriateness of the fatty liver inhibition of progression (FLIP) algorithm and steatosis, activity, and fibrosis (SAF) score in the evaluation of biopsies of nonalcoholic fatty liver disease. Hepatology 2014, 60, 565–575. [Google Scholar] [CrossRef]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef] [Green Version]
- Stine, J.G.; Wentworth, B.J.; Zimmet, A.; Rinella, M.E.; Loomba, R.; Caldwell, S.H.; Argo, C.K. Systematic review with meta-analysis: Risk of hepatocellular carcinoma in non-alcoholic steatohepatitis without cirrhosis compared to other liver diseases. Aliment. Pharmacol. Ther. 2018, 48, 696–703. [Google Scholar] [CrossRef]
- Ioannou, G.N.; Splan, M.F.; Weiss, N.S.; McDonald, G.B.; Beretta, L.; Lee, S.P. Incidence and predictors of hepatocellular carcinoma in patients with cirrhosis. Clin. Gastroenterol. Hepatol. 2007, 5, 938–945.e934. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease—Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Peng, C.; Stewart, A.G.; Woodman, O.L.; Ritchie, R.H.; Qin, C.X. Non-alcoholic steatohepatitis: A review of its mechanism, models and medical treatments. Front. Pharmacol. 2020, 11, 603926. [Google Scholar] [CrossRef] [PubMed]
- Hasin-Brumshtein, Y.; Sakaram, S.; Khatri, P.; He, Y.D.; Sweeney, T.E. A robust gene expression signature for NASH in liver expression data. Sci. Rep. 2022, 12, 2571. [Google Scholar] [CrossRef] [PubMed]
- Trichopoulou, A.; Costacou, T.; Bamia, C.; Trichopoulos, D. Adherence to a Mediterranean diet and survival in a Greek population. N. Engl. J. Med. 2003, 348, 2599–2608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keys, A. Seven countries. In Seven Countries; Harvard University Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Estruch, R.; Ros, E.; Salas-Salvado, J.; Covas, M.I.; Corella, D.; Aros, F.; Gomez-Gracia, E.; Ruiz-Gutierrez, V.; Fiol, M.; Lapetra, J.; et al. Primary Prevention of Cardiovascular Disease with a Mediterranean Diet Supplemented with Extra-Virgin Olive Oil or Nuts. N. Engl. J. Med. 2018, 378, e34. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Marcos, L.V.; Lou-Bonafonte, J.M.; Arnal, C.; Navarro, M.A.; Osada, J. Transcriptomics and the Mediterranean Diet: A Systematic Review. Nutrients 2017, 9, 472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Beamonte, R.; Sanclemente, T.; Surra, J.C.; Osada, J. Could squalene be an added value to use olive by-products? J. Sci. Food Agric. 2020, 100, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Tomkins, G.; Dauben, W.; Sheppard, H.; Chaikoff, I. Squalene as a precursor of cholesterol in liver. J. Biol. Chem. 1953, 202, 487–489. [Google Scholar] [CrossRef]
- Lou-Bonafonte, J.M.; Martinez-Beamonte, R.; Sanclemente, T.; Surra, J.C.; Herrera-Marcos, L.V.; Sanchez-Marco, J.; Arnal, C.; Osada, J. Current Insights into the Biological Action of Squalene. Mol. Nutr. Food Res. 2018, 8, e1800136. [Google Scholar] [CrossRef]
- Martinez-Beamonte, R.; Sanchez-Marco, J.; Felices, M.J.; Barranquero, C.; Gascon, S.; Arnal, C.; Burillo, J.C.; Lasheras, R.; Busto, R.; Lasuncion, M.A.; et al. Dietary squalene modifies plasma lipoproteins and hepatic cholesterol metabolism in rabbits. Food Funct. 2021, 12, 8141–8153. [Google Scholar] [CrossRef]
- Tsujimoto, M. A highly unsaturated hydrocarbon in shark liver oil. J. Ind. Eng. Chem. 1916, 8, 889–896. [Google Scholar] [CrossRef] [Green Version]
- Wetherbee, B.M.; Nichols, P.D. Lipid composition of the liver oil of deep-sea sharks from the Chatham Rise, New Zealand. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2000, 125, 511–521. [Google Scholar] [CrossRef]
- Nielsen, J.; Hedeholm, R.B.; Heinemeier, J.; Bushnell, P.G.; Christiansen, J.S.; Olsen, J.; Ramsey, C.B.; Brill, R.W.; Simon, M.; Steffensen, K.F.; et al. Eye lens radiocarbon reveals centuries of longevity in the Greenland shark (Somniosus microcephalus). Science 2016, 353, 702–704. [Google Scholar] [CrossRef]
- MacNeil, M.A.; McMeans, B.C.; Hussey, N.E.; Vecsei, P.; Svavarsson, J.; Kovacs, K.M.; Lydersen, C.; Treble, M.A.; Skomal, G.B.; Ramsey, M.; et al. Biology of the Greenland shark Somniosus microcephalus. J. Fish. Biol. 2012, 80, 991–1018. [Google Scholar] [CrossRef]
- Kritchevsky, D.; Moyer, A.W.; Tesar, W.C.; Logan, J.B.; Brown, R.A.; Richmond, G. Squalene feeding in experimental atherosclerosis. Circ. Res. 1954, 2, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Beamonte, R.; Alda, O.; Sanclemente, T.; Felices, M.J.; Escusol, S.; Arnal, C.; Herrera-Marcos, L.V.; Gascon, S.; Surra, J.C.; Osada, J.; et al. Hepatic subcellular distribution of squalene changes according to the experimental setting. J. Physiol. Biochem. 2018, 74, 531–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillen, N.; Acin, S.; Navarro, M.A.; Perona, J.S.; Arbones-Mainar, J.M.; Arnal, C.; Sarria, A.J.; Surra, J.C.; Carnicer, R.; Orman, I.; et al. Squalene in a sex-dependent manner modulates atherosclerotic lesion which correlates with hepatic fat content in apoE-knockout male mice. Atherosclerosis 2008, 197, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Torres, A.; Barcelo-Batllori, S.; Martinez-Beamonte, R.; Navarro, M.A.; Surra, J.C.; Arnal, C.; Guillen, N.; Acin, S.; Osada, J. Proteomics and gene expression analyses of squalene-supplemented mice identify microsomal thioredoxin domain-containing protein 5 changes associated with hepatic steatosis. J. Proteom. 2012, 77, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Torres, A.; Barcelo-Batllori, S.; Fernandez-Vizarra, E.; Navarro, M.A.; Arnal, C.; Guillen, N.; Acin, S.; Osada, J. Proteomics and gene expression analyses of mitochondria from squalene-treated apoE-deficient mice identify short-chain specific acyl-CoA dehydrogenase changes associated with fatty liver amelioration. J. Proteom. 2012, 75, 2563–2575. [Google Scholar] [CrossRef]
- Gabas-Rivera, C.; Jurado-Ruiz, E.; Sanchez-Ortiz, A.; Romanos, E.; Martinez-Beamonte, R.; Navarro, M.A.; Surra, J.C.; Arnal, C.; Rodriguez-Yoldi, M.J.; Andres-Lacueva, C.; et al. Dietary Squalene Induces Cytochromes Cyp2b10 and Cyp2c55 Independently of Sex, Dose, and Diet in Several Mouse Models. Mol. Nutr. Food Res. 2020, 64, e2000354. [Google Scholar] [CrossRef]
- Abuobeid, R.; Sánchez-Marco, J.; Felices, M.J.; Arnal, C.; Burillo, J.C.; Lasheras, R.; Busto, R.; Lasunción, M.A.; Rodríguez-Yoldi, M.J.; Martínez-Beamonte, R. Squalene through Its Post-Squalene Metabolites Is a Modulator of Hepatic Transcriptome in Rabbits. Int. J. Mol. Sci. 2022, 23, 4172. [Google Scholar] [CrossRef] [PubMed]
- Bassols, A.; Costa, C.; Eckersall, P.D.; Osada, J.; Sabria, J.; Tibau, J. The pig as an animal model for human pathologies: A proteomics perspective. Proteom. Clin. Appl. 2014, 8, 715–731. [Google Scholar] [CrossRef]
- Herrera-Marcos, L.V.; Martinez-Beamonte, R.; Macias-Herranz, M.; Arnal, C.; Barranquero, C.; Puente-Lanzarote, J.J.; Gascon, S.; Herrero-Continente, T.; Gonzalo-Romeo, G.; Alastrue-Vera, V.; et al. Hepatic galectin-3 is associated with lipid droplet area in non-alcoholic steatohepatitis in a new swine model. Sci. Rep. 2022, 12, 1024. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Marcos, L.V.; Martínez-Beamonte, R.; Arnal, C.; Barranquero, C.; Puente-Lanzarote, J.J.; Herrero-Continente, T.; Lou-Bonafonte, J.M.; Gonzalo-Romeo, G.; Mocciaro, G.; Jenkins, B. Dietary squalene supplementation decreases triglyceride species and modifies phospholipid lipidomic profile in the liver of a porcine model of non-alcoholic steatohepatitis. J. Nutr. Biochem. 2023, 112, 109207. [Google Scholar] [CrossRef] [PubMed]
- Bidooki, S.H.; Alejo, T.; Sánchez-Marco, J.; Martínez-Beamonte, R.; Abuobeid, R.; Burillo, J.C.; Lasheras, R.; Sebastian, V.; Rodríguez-Yoldi, M.J.; Arruebo, M. Squalene Loaded Nanoparticles Effectively Protect Hepatic AML12 Cell Lines against Oxidative and Endoplasmic Reticulum Stress in a TXNDC5-Dependent Way. Antioxidants 2022, 11, 581. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, A.P.; Qvist, P.; Lazarus, R.; Lescai, F.; Ju, J.; Nyegaard, M.; Mors, O.; Børglum, A.D.; Li, Q.; Christensen, J.H. Experimental validation of methods for differential gene expression analysis and sample pooling in RNA-seq. BMC Genom. 2015, 16, 548. [Google Scholar] [CrossRef] [Green Version]
- Sham, P.; Bader, J.S.; Craig, I.; O’Donovan, M.; Owen, M. DNA pooling: A tool for large-scale association studies. Nat. Rev. Genet. 2002, 3, 862–871. [Google Scholar] [CrossRef]
- Peng, X.; Wood, C.L.; Blalock, E.M.; Chen, K.C.; Landfield, P.W.; Stromberg, A.J. Statistical implications of pooling RNA samples for microarray experiments. BMC Bioinform. 2003, 4, 26. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Fukusato, T. Animal models of liver diseases. In Animal Models for the Study of Human Disease; Elsevier: Amsterdam, The Netherlands, 2017; pp. 313–339. [Google Scholar]
- Muriel, P.; Ramos-Tovar, E.; Montes-Páez, G.; Buendía-Montaño, L. Experimental models of liver damage mediated by oxidative stress. In Liver Pathophysiology; Elsevier: Amsterdam, The Netherlands, 2017; pp. 529–546. [Google Scholar]
- Gabas-Rivera, C.; Barranquero, C.; Martinez-Beamonte, R.; Navarro, M.A.; Surra, J.C.; Osada, J. Dietary squalene increases high density lipoprotein-cholesterol and paraoxonase 1 and decreases oxidative stress in mice. PLoS ONE 2015, 9, e104224. [Google Scholar] [CrossRef]
- Prather, R.S.; Lorson, M.; Ross, J.W.; Whyte, J.J.; Walters, E. Genetically engineered pig models for human diseases. Annu. Rev. Anim. Biosci. 2013, 1, 203. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, M.; Sjöstedt, E.; Oksvold, P.; Sivertsson, Å.; Huang, J.; Álvez, M.B.; Arif, M.; Li, X.; Lin, L.; Yu, J. Genome-wide annotation of protein-coding genes in pig. BMC Biol. 2022, 20, 25. [Google Scholar] [CrossRef]
- Hoang, T.M.H.; Nguyen, C.H.; Le, T.T.; Hoang, T.H.Q.; Ngo, T.H.T.; Hoang, T.L.A.; Dang, D.H. Squalene isolated from Schizochytrium mangrovei is a peroxisome proliferator-activated receptor-α agonist that regulates lipid metabolism in HepG2 cells. Biotechnol. Lett. 2016, 38, 1065–1071. [Google Scholar] [CrossRef]
- Niu, L.; Geyer, P.E.; Wewer Albrechtsen, N.J.; Gluud, L.L.; Santos, A.; Doll, S.; Treit, P.V.; Holst, J.J.; Knop, F.K.; Vilsbøll, T. Plasma proteome profiling discovers novel proteins associated with non-alcoholic fatty liver disease. Mol. Syst. Biol. 2019, 15, e8793. [Google Scholar] [CrossRef]
- Raza-Iqbal, S.; Tanaka, T.; Anai, M.; Inagaki, T.; Matsumura, Y.; Ikeda, K.; Taguchi, A.; Gonzalez, F.J.; Sakai, J.; Kodama, T. Transcriptome analysis of K-877 (a novel selective PPARα modulator (SPPARMα))-regulated genes in primary human hepatocytes and the mouse liver. J. Atheroscler. Thromb. 2015, 22, 754–772. [Google Scholar] [CrossRef] [Green Version]
- Fruchart, J.-C. Pemafibrate (K-877), a novel selective peroxisome proliferator-activated receptor alpha modulator for management of atherogenic dyslipidaemia. Cardiovasc. Diabetol. 2017, 16, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florentin, M.; Kostapanos, M.S.; Anagnostis, P.; Liamis, G. Recent developments in pharmacotherapy for hypertriglyceridemia: What’s the current state of the art? Expert. Opin. Pharmacother. 2020, 21, 107–120. [Google Scholar] [CrossRef]
- Rosenblat, M.; Volkova, N.; Aviram, M. Pomegranate juice (PJ) consumption antioxidative properties on mouse macrophages, but not PJ beneficial effects on macrophage cholesterol and triglyceride metabolism, are mediated via PJ-induced stimulation of macrophage PON2. Atherosclerosis 2010, 212, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Cichoż-Lach, H.; Michalak, A. Oxidative stress as a crucial factor in liver diseases. World J. Gastroenterol. WJG 2014, 20, 8082. [Google Scholar] [CrossRef]
- Lee, J.; Homma, T.; Kurahashi, T.; Kang, E.S.; Fujii, J. Oxidative stress triggers lipid droplet accumulation in primary cultured hepatocytes by activating fatty acid synthesis. Biochem. Biophys. Res. Commun. 2015, 464, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Malott, K.F.; Reshel, S.; Ortiz, L.; Luderer, U. Glutathione deficiency decreases lipid droplet stores and increases reactive oxygen species in mouse oocytes. Biol. Reprod. 2022, 106, 1218–1231. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, T.J.; Vargas-Guerrero, B.; García-López, P.M.; Gurrola-Díaz, C.M. Analysis of hepatic transcriptome modulation exerted by γ-conglutin from lupins in a streptozotocin-induced diabetes model. Gene 2020, 761, 145036. [Google Scholar] [CrossRef] [PubMed]
- Parhofer, K.G. Interaction between glucose and lipid metabolism: More than diabetic dyslipidemia. Diabetes Metab. J. 2015, 39, 353–362. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Liu, Y.; Qian, Y.; Shen, Z.; He, Y.; Gao, R.; Shen, M.; Chen, S.; Fu, Q.; Yang, T. CHL1 promotes insulin secretion and negatively regulates the proliferation of pancreatic β cells. Biochem. Biophys. Res. Commun. 2020, 525, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Taneera, J.; Dhaiban, S.; Hachim, M.; Mohammed, A.K.; Mukhopadhyay, D.; Bajbouj, K.; Hamoudi, R.; Salehi, A.; Hamad, M. Reduced expression of Chl1 gene impairs insulin secretion by down-regulating the expression of key molecules of β-cell function. Exp. Clin. Endocrinol. Diabetes 2021, 129, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Chirieac, D.V.; Chirieac, L.R.; Corsetti, J.P.; Cianci, J.; Sparks, C.E.; Sparks, J.D. Glucose-stimulated insulin secretion suppresses hepatic triglyceride-rich lipoprotein and apoB production. Am. J. Physiol. Endocrinol. Metab. 2000, 279, E1003–E1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adiels, M.; Taskinen, M.-R.; Borén, J. Fatty liver, insulin resistance, and dyslipidemia. Curr. Diabetes Rep. 2008, 8, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Tricò, D.; Natali, A.; Mari, A.; Ferrannini, E.; Santoro, N.; Caprio, S. Triglyceride-rich very low-density lipoproteins (VLDL) are independently associated with insulin secretion in a multiethnic cohort of adolescents. Diabetes Obes. Metab. 2018, 20, 2905–2910. [Google Scholar] [CrossRef]
- Khan, A.; Molitor, A.; Mayeur, S.; Zhang, G.; Rinaldi, B.; Lannes, B.; Lhermitte, B.; Umair, M.; Arold, S.T.; Friant, S. A Homozygous Missense Variant in PPP1R1B/DARPP-32 Is Associated With Generalized Complex Dystonia. Mov. Disord. 2022, 37, 365–374. [Google Scholar] [CrossRef]
- Brady, M.J.; Saltiel, A.R. The role of protein phosphatase-1 in insulin action. Recent. Prog. Horm. Res. 2001, 56, 157–174. [Google Scholar] [CrossRef]
- Allende, D.S.; Gawrieh, S.; Cummings, O.W.; Belt, P.; Wilson, L.; Van Natta, M.; Behling, C.A.; Carpenter, D.; Gill, R.M.; Kleiner, D.E. Glycogenosis is common in nonalcoholic fatty liver disease and is independently associated with ballooning, but lower steatosis and lower fibrosis. Liver Int. 2021, 41, 996–1011. [Google Scholar] [CrossRef]
- Yamada, K.; Mizukoshi, E.; Sunagozaka, H.; Arai, K.; Yamashita, T.; Takeshita, Y.; Misu, H.; Takamura, T.; Kitamura, S.; Zen, Y. Characteristics of hepatic fatty acid compositions in patients with nonalcoholic steatohepatitis. Liver Int. 2015, 35, 582–590. [Google Scholar] [CrossRef] [Green Version]
- Yoo, W.; Gjuka, D.; Stevenson, H.L.; Song, X.; Shen, H.; Yoo, S.Y.; Wang, J.; Fallon, M.; Ioannou, G.N.; Harrison, S.A. Fatty acids in non-alcoholic steatohepatitis: Focus on pentadecanoic acid. PLoS ONE 2017, 12, e0189965. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Liu, C.; Zhou, D.; Zhang, L. TGF-β/SMAD pathway and its regulation in hepatic fibrosis. J. Histochem. Cytochem. 2016, 64, 157–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Lin, W.; Fang, P.; Lin, X.; Zhuge, L.; Hu, Z.; Jin, L. MiR-10a improves hepatic fibrosis by regulating the TGFβl/Smads signal transduction pathway. Exp. Ther. Med. 2016, 12, 1719–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arimoto, K.I.; Miyauchi, S.; Stoner, S.A.; Fan, J.B.; Zhang, D.E. Negative regulation of type I IFN signaling. J. Leukoc. Biol. 2018, 103, 1099–1116. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, S.; Yang, M. The Role of Interferon Regulatory Factors in Non-Alcoholic Fatty Liver Disease and Non-Alcoholic Steatohepatitis. Gastroenterol. Insights 2022, 13, 16. [Google Scholar] [CrossRef]
- Kersten, S.; Stienstra, R. The role and regulation of the peroxisome proliferator activated receptor alpha in human liver. Biochimie 2017, 136, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Tan, J.; Dong, W.; Zou, B.; Teng, X.; Zhu, L.; Ge, C.; Dai, X.; Kuang, Q.; Zhong, S. The E3 ubiquitin-protein ligase Trim31 alleviates non-alcoholic fatty liver disease by targeting Rhbdf2 in mouse hepatocytes. Nat. Commun. 2022, 13, 1052. [Google Scholar] [CrossRef]
- Gawrieh, S.; Baye, T.M.; Carless, M.; Wallace, J.; Komorowski, R.; Kleiner, D.E.; Andris, D.; Makladi, B.; Cole, R.; Charlton, M. Hepatic gene networks in morbidly obese patients with nonalcoholic fatty liver disease. Obes. Surg. 2010, 20, 1698–1709. [Google Scholar] [CrossRef]
- Smith, T.J. Squalene: Potential chemopreventive agent. Expert. Opin. Investig. Drugs 2000, 9, 1841–1848. [Google Scholar] [CrossRef]
- van Huizen, N.A.; van den Braak, R.R.C.; Doukas, M.; Dekker, L.J.; IJzermans, J.N.; Luider, T.M. Up-regulation of collagen proteins in colorectal liver metastasis compared with normal liver tissue. J. Biol. Chem. 2019, 294, 281–289. [Google Scholar] [CrossRef] [Green Version]
- Sirivatanauksorn, Y.; Sirivatanauksorn, V.; Srisawat, C.; Khongmanee, A.; Tongkham, C. Differential expression of sprouty genes in hepatocellular carcinoma. J. Surg. Oncol. 2012, 105, 273–276. [Google Scholar] [CrossRef]
- Ghosheh, N.; Küppers-Munther, B.; Asplund, A.; Edsbagge, J.; Ulfenborg, B.; Andersson, T.B.; Björquist, P.; Andersson, C.X.; Carén, H.; Simonsson, S. Comparative transcriptomics of hepatic differentiation of human pluripotent stem cells and adult human liver tissue. Physiol. Genom. 2017, 49, 430–446. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.-M.; Wu, T.-H.; Wu, T.-J.; Chou, H.-S.; Yu, M.-C.; Lee, W.-C. Bioinformatics microarray analysis and identification of gene expression profiles associated with cirrhotic liver. Kaohsiung J. Med. Sci. 2016, 32, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.; Hu, F.; Xie, X.; Wang, L.; Li, G.; Qiao, T.; Wang, M.; Xiao, H. TMEM45B, up-regulated in human lung cancer, enhances tumorigenicity of lung cancer cells. Tumor Biol. 2016, 37, 12181–12191. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.-c.; Shen, B.-y.; Deng, X.-x.; Chen, H.; Zhu, Z.-g.; Peng, C.-h. TMEM45B promotes proliferation, invasion and migration and inhibits apoptosis in pancreatic cancer cells. Mol. BioSyst. 2016, 12, 1860–1870. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Yu, W.; Yu, Y.; Liu, X.; Cui, X. Knockdown of TMEM45B inhibits cell proliferation and invasion in gastric cancer. Biomed. Pharmacother. 2018, 104, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Guo, S.; Zhang, W.; Li, D.; Wang, Y.; Zhan, Q. Co-expression network analysis identifies key modules and hub genes implicated in esophageal squamous cell cancer progression. Med. Omics 2021, 1, 100003. [Google Scholar] [CrossRef]
- Senchenko, V.N.; Krasnov, G.S.; Dmitriev, A.A.; Kudryavtseva, A.V.; Anedchenko, E.A.; Braga, E.A.; Pronina, I.V.; Kondratieva, T.T.; Ivanov, S.V.; Zabarovsky, E.R. Differential expression of CHL1 gene during development of major human cancers. PLoS ONE 2011, 6, e15612. [Google Scholar] [CrossRef]
- Abuobeid, R.; Herrera-Marcos, L.; Navarro, M.A.; Arnal, C.; Martinez-Beamonte, R.; Surra, J.; Osada, J. Dietary Erythrodiol Modifies Hepatic Transcriptome in Mice in a Sex and Dose-Dependent Way. Int. J. Mol. Sci. 2020, 21, 21197331. [Google Scholar] [CrossRef]







| Function | Gene ID | Name | Symbol | Control (FPKM) | Squalene (FPKM) | Log2 Fold Change Squalene/Control |
|---|---|---|---|---|---|---|
| Up-regulated | ||||||
| Signal transduction | 100736966 | Protein phosphatase 1 regulatory inhibitor subunit 1B | PPP1R1B | 63 | 785 | 3.7 |
| Signal transduction | 595119 | 2′-5′-oligoadenylate synthase-like protein | OASL | 441 | 3611 | 3.0 |
| Signal transduction | 100737576 | Protein phosphatase 4 regulatory subunit 4 | PPP4R4 | 59 | 314 | 2.4 |
| Transcription factor | 100739264 | Hes family bHLH transcription factor 4 | HES4 | 569 | 2853 | 2.3 |
| Protein ubiquinization | 100524797 | Neuralized E3 ubiquitin protein ligase 3 | NEURL3 | 98 | 456 | 2.2 |
| Fatty acid biosynthesis | 100515579 | Hydroxyacyl-thioester dehydratase type 2, mitochondrial | LOC100515579 (HTD2) | 259 | 1155 | 2.2 |
| Metabolism of xenobiotics | 403106 | Cytochrome P450, 2C32 | CYP2C32 | 2165 | 8633 | 2.0 |
| Membrane protein | 100516991 | Transmembrane protein 45B | TMEM45B | 237 | 825 | 1.8 |
| Fatty acid transport | 397586 | Alpha-fetoprotein | AFP | 77 | 268 | 1.8 |
| Peptide hormone metabolism | 397080 | Glutamyl aminopeptidase | ENPEP | 191 | 647 | 1.8 |
| Endogenous retrovirus C | 110256649 | Porcine endogenous retrovirus C gag protein | LOC110256649 | 337 | 1071 | 1.7 |
| Metabolism of xenobiotics | 100524750 | Cytochrome P450, 2J34 | CYP2J34 | 414 | 1249 | 1.6 |
| Metabolism of xenobiotics | 100526118 | Glutathione S-transferase A1-like | LOC100526118 | 5125 | 15029 | 1.6 |
| Cell cycle progression | 100152729 | S100 calcium binding protein A2 | S100A2 | 1817 | 5171 | 1.5 |
| ERK signaling | 100515521 | Sprouty RTK signaling antagonist 3 | SPRY3 | 301 | 848 | 1.5 |
| Transcription factor | 110261579 | Forkhead box G1 | FOXG1 | 134 | 376 | 1.5 |
| Down-regulated | ||||||
| Control of transcription | 100521495 | Gametocyte specific factor 1 | GTSF1 | 543 | 182 | −1.6 |
| Sterol biosynthesis | 100113409 | Squalene monooxygenase | SQLE | 464 | 112 | −2.0 |
| Signal transduction | 100511780 | Cell adhesion molecule L1 like | CHL1 | 570 | 34 | −4.1 |
| Gene Symbol | Control (n = 12) | Squalene (n = 12) | Fold Change | Signal log2 Ratio (SL2R) |
|---|---|---|---|---|
| PPP1R1B | 2.2 ± 3.1 | 9.0 ± 10.5 * | 4.2 | 2.1 |
| OASL | 0.8 ± 0.7 | 2.5 ± 1.4 * | 3.1 | 1.6 |
| PPP4R4 | 1.1 ± 0.8 | 6.7 ± 13.2 * | 6.1 | 2.6 |
| HES4 | 1.3 ± 1.3 | 7.2 ± 20.2 | 5.4 | 2.4 |
| NEURL3 | 1.8 ± 2.2 | 11.3 ± 23.6 * | 6.2 | 2.6 |
| LOC100515579 (HTD2) | 1.1 ± 0.4 | 1.1 ± 0.8 | 1.0 | 0.05 |
| CYP2C32 | 2.8 ± 4.7 | 15.2 ± 24.6 | 5.3 | 2.4 |
| TMEM45B | 2.5 ± 4.7 | 11.1 ± 15.8 * | 4.4 | 2.1 |
| AFP | 1.3 ± 0.9 | 5.4 ± 8.5 ** | 4.1 | 2.0 |
| ENPEP | 1.3 ± 0.7 | 6.3 ± 9.7 ** | 4.9 | 2.3 |
| LOC110256649 | 1.6 ± 1.7 | 3.7 ± 3.2 * | 2.4 | 1.2 |
| CYP2J34 | 2.0 ± 1.6 | 4.9 ± 9.6 | 2.5 | 1.3 |
| LOC100526118 | 1.4 ± 1.6 | 3.5 ± 4.8 ** | 2.5 | 1.3 |
| S100A2 | 1.9 ± 1.9 | 4.4 ± 7.0 | 2.3 | 1.2 |
| SPRY3 | 1.2 ± 0.7 | 4.3 ± 5.2 * | 3.5 | 1.8 |
| FOXG1 | 1.9 ± 2.2 | 6.8 ± 11.7 | 3.7 | 1.9 |
| GTSF1 | 4.0 ± 5.7 | 1.7 ± 1.7 | 0.4 | −1.2 |
| SQLE | 5.1 ± 14.5 | 2.7 ± 1.9 * | 0.5 | −0.9 |
| CHL1 | 6.5 ± 11.9 | 0.2 ± 0.2 * | 0.03 | −4.8 |
| Gene Symbol | Control (n = 6) | Squalene (n = 6) | Fold Change | Signal log2 Ratio |
|---|---|---|---|---|
| PPP1R1B | 0.7 ± 0.2 | 0.9 ± 0.2 * | 1.3 | 0.4 |
| TMEM45B | 1.0 ± 0.1 | 1.3 ± 0.1 ** | 1.3 | 0.4 |
| AFP | 0.8 ± 0.2 | 1.0 ± 0.1 * | 1.2 | 0.2 |
| ENPEP | 0.8 ± 0.2 | 1.5 ± 0.3 * | 1.9 | 0.9 |
| SPRY3 | 0.8 ± 0.3 | 1.4 ± 0.3 * | 1.8 | 0.8 |
| Gene Symbol | Control (n = 6) | Squalene (n = 6) | Fold Change | Signal log2 Ratio |
|---|---|---|---|---|
| Ppp1r1b | 1.0 ± 0.1 | 1.5 ± 0.5 * | 1.5 | 0.6 |
| Tmem45b | 1.0 ± 0.3 | 3.0 ± 1.7 * | 3.0 | 1.6 |
| Afp | 1.0 ± 0.1 | 1.6 ± 0.8 * | 1.6 | 0.7 |
| Enpep | 1.0 ± 0.1 | 1.4 ± 0.5 * | 1.4 | 0.5 |
| Spry3 | 1.2 ± 0.1 | 0.7 ± 0.1 * | 0.7 | −0.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abuobeid, R.; Herrera-Marcos, L.V.; Arnal, C.; Bidooki, S.H.; Sánchez-Marco, J.; Lasheras, R.; Surra, J.C.; Rodríguez-Yoldi, M.J.; Martínez-Beamonte, R.; Osada, J. Differentially Expressed Genes in Response to a Squalene-Supplemented Diet Are Accurate Discriminants of Porcine Non-Alcoholic Steatohepatitis. Int. J. Mol. Sci. 2023, 24, 12552. https://doi.org/10.3390/ijms241612552
Abuobeid R, Herrera-Marcos LV, Arnal C, Bidooki SH, Sánchez-Marco J, Lasheras R, Surra JC, Rodríguez-Yoldi MJ, Martínez-Beamonte R, Osada J. Differentially Expressed Genes in Response to a Squalene-Supplemented Diet Are Accurate Discriminants of Porcine Non-Alcoholic Steatohepatitis. International Journal of Molecular Sciences. 2023; 24(16):12552. https://doi.org/10.3390/ijms241612552
Chicago/Turabian StyleAbuobeid, Roubi, Luis V. Herrera-Marcos, Carmen Arnal, Seyed Hesamoddin Bidooki, Javier Sánchez-Marco, Roberto Lasheras, Joaquín C. Surra, María Jesús Rodríguez-Yoldi, Roberto Martínez-Beamonte, and Jesús Osada. 2023. "Differentially Expressed Genes in Response to a Squalene-Supplemented Diet Are Accurate Discriminants of Porcine Non-Alcoholic Steatohepatitis" International Journal of Molecular Sciences 24, no. 16: 12552. https://doi.org/10.3390/ijms241612552