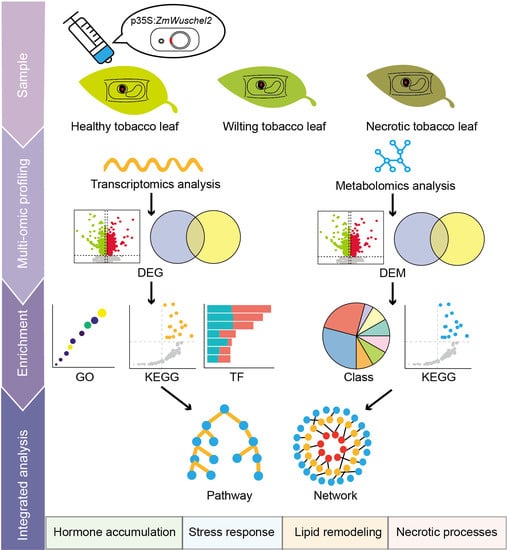
Transcriptomic and Metabolomic Investigation on Leaf Necrosis Induced by ZmWus2 Transient Overexpression in Nicotiana benthamiana
Abstract
:1. Introduction
2. Results
2.1. Transient Expression of ZmWus2 Leads to Tobacco Leaf Necrosis
2.2. Transcriptome Changes in Tobacco Leaves Induced by ZmWus2 Transfection
2.3. Biological Insights through Transcriptomic DEGs
2.4. Metabolomic Changes in Tobacco Leaves Induced by ZmWus2 Transfection
2.5. Functional Annotation of Metabolomic DEMs
2.6. Metabolic Pathways Regulated by ZmWus2
2.7. Regulated Metabolic Pathways Evident in Both Transcriptome and Metabolome
2.8. Transcription Factor Regulated by ZmWus2
3. Discussion
4. Materials and Methods
4.1. Construction of a Binary Vector for Tobacco Transformation
4.2. Agroinfiltration of Tobacco Leaves
4.3. Subcellular Localization
4.4. Ultra-Thin Section and Semi-Thin Section
4.5. Transcriptomic Analysis
4.6. Metabolomic Analysis
4.7. Data Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gallois, J.L.; Nora, F.R.; Mizukami, Y.; Sablowski, R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 2004, 18, 375–380. [Google Scholar] [CrossRef] [Green Version]
- Jha, P.; Ochatt, S.J.; Kumar, V. WUSCHEL: A master regulator in plant growth signaling. Plant Cell Rep. 2020, 39, 431–444. [Google Scholar] [CrossRef]
- Lopes, F.-C.L.; Galvan-Ampudia, C.; Landrein, B. WUSCHEL in the shoot apical meristem: Old player, new tricks. J. Exp. Bot. 2020, 72, 1527–1535. [Google Scholar] [CrossRef]
- Somssich, M.; Je, B.I.; Simon, R.; Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016, 143, 3238–3248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jönsson, H.; Reddy, G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011, 25, 2025–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leibfried, A.; To, J.P.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 2005, 438, 1172–1175. [Google Scholar] [CrossRef]
- Ma, Y.; Miotk, A.; Šutiković, Z.; Ermakova, O.; Wenzl, C.; Medzihradszky, A.; Gaillochet, C.; Forner, J.; Utan, G.; Brackmann, K.; et al. WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat. Commun. 2019, 10, 5093. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.-Q.; Lian, H.; Zhou, C.-M.; Xu, L.; Jiao, Y.; Wang, J.-W. A Two-Step Model for de Novo Activation of WUSCHEL during Plant Shoot Regeneration. Plant Cell 2017, 29, 1073–1087. [Google Scholar] [CrossRef] [Green Version]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P.; et al. Advancing Crop Transformation in the Era of Genome Editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, J.; Niu, Q.-W.; Frugis, G.; Chua, N.-H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002, 30, 349–359. [Google Scholar] [CrossRef]
- Gordon-Kamm, B.; Sardesai, N.; Arling, M.; Lowe, K.; Hoerster, G.; Betts, S.; Jones, T. Using Morphogenic Genes to Improve Recovery and Regeneration of Transgenic Plants. Plants 2019, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, P.; Wu, E.; Simon, M.K.; Anand, A.; Lowe, K.; Gao, H.; Sigmund, A.L.; Yang, M.; Albertsen, M.C.; Gordon-Kamm, W.; et al. Wuschel2 enables highly efficient CRISPR/Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. Commun. Biol. 2022, 5, 344. [Google Scholar] [CrossRef]
- Bouchabké-Coussa, O.; Obellianne, M.; Linderme, D.; Montes, E.; Maia-Grondard, A.; Vilaine, F.; Pannetier, C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013, 32, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Lowe, K.; La Rota, M.; Hoerster, G.; Hastings, C.; Wang, N.; Chamberlin, M.; Wu, E.; Jones, T.; Gordon-Kamm, W. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev. Biol. Plant 2018, 54, 240–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maher, M.F.; Nasti, R.A.; Vollbrecht, M.; Starker, C.G.; Clark, M.D.; Voytas, D.F. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020, 38, 84–89. [Google Scholar] [CrossRef]
- Fambrini, M.; Usai, G.; Pugliesi, C. Induction of Somatic Embryogenesis in Plants: Different Players and Focus on WUSCHEL and WUS-RELATED HOMEOBOX (WOX) Transcription Factors. Int. J. Mol. Sci. 2022, 23, 15950. [Google Scholar] [CrossRef]
- Drøbak, B.K.; Watkins, P.A. Inositol(1,4,5)trisphosphate production in plant cells: An early response to salinity and hyperosmotic stress. FEBS Lett. 2000, 481, 240–244. [Google Scholar] [CrossRef]
- Jia, Q.; Kong, D.; Li, Q.; Sun, S.; Song, J.; Zhu, Y.; Liang, K.; Ke, Q.; Lin, W.; Huang, J. The Function of Inositol Phosphatases in Plant Tolerance to Abiotic Stress. Int. J. Mol. Sci. 2019, 20, 3999. [Google Scholar] [CrossRef] [Green Version]
- Tuteja, N.; Ahmad, P.; Panda, B.B.; Tuteja, R. Genotoxic stress in plants: Shedding light on DNA damage, repair and DNA repair helicases. Mutat. Res. Mol. Mech. Mutagen. 2009, 681, 134–149. [Google Scholar] [CrossRef]
- Awada, R.; Campa, C.; Gibault, E.; Déchamp, E.; Georget, F.; Lepelley, M.; Abdallah, C.; Erban, A.; Martinez-Seidel, F.; Kopka, J.; et al. Unravelling the Metabolic and Hormonal Machinery During Key Steps of Somatic Embryogenesis: A Case Study in Coffee. Int. J. Mol. Sci. 2019, 20, 4665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, F.; Schon, M.A.; Nodine, M.D. The embryonic transcriptome of Arabidopsis thaliana. Plant Reprod. 2019, 32, 77–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awada, R.; Lepelley, M.; Breton, D.; Charpagne, A.; Campa, C.; Berry, V.; Georget, F.; Breitler, J.-C.; Léran, S.; Djerrab, D.; et al. Global transcriptome profiling reveals differential regulatory, metabolic and hormonal networks during somatic embryogenesis in Coffea arabica. BMC Genom. 2023, 24, 41. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Raza, A.; Gao, A.; Zeng, L.; Lv, Y.; Ding, X.; Cheng, Y.; Zou, X. Plant lipid phosphate phosphatases: Current advances and future outlooks. Crit. Rev. Biotechnol. 2023, 43, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.-J.; Reddy, M.S.S.; Wang, L. The phenylpropanoid pathway and plant defence-a genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef]
- Olivares-García, C.A.; Mata-Rosas, M.; Peña-Montes, C.; Quiroz-Figueroa, F.; Segura-Cabrera, A.; Shannon, L.M.; Loyola-Vargas, V.M.; Monribot-Villanueva, J.L.; Elizalde-Contreras, J.M.; Ibarra-Laclette, E.; et al. Phenylpropanoids Are Connected to Cell Wall Fortification and Stress Tolerance in Avocado Somatic Embryogenesis. Int. J. Mol. Sci. 2020, 21, 5679. [Google Scholar] [CrossRef]
- Ameri, A.; Davarynejad, G.H.; Moshtaghi, N.; Tehranifar, A. The Role of Carbohydrates on The Induction of Somatic Embryogenesis and The Biochemical State of The Embryogenic Callus in Pyrus communis L. Cv. ‘Dar Gazi’. Erwerbs-Obstbau 2020, 62, 411–419. [Google Scholar] [CrossRef]
- Hofhuis, H.F.; Heidstra, R. Transcription factor dosage: More or less sufficient for growth. Curr. Opin. Plant Biol. 2018, 45, 50–58. [Google Scholar] [CrossRef]
- Gliwicka, M.; Nowak, K.; Balazadeh, S.; Mueller-Roeber, B.; Gaj, M.D. Extensive Modulation of the Transcription Factor Transcriptome during Somatic Embryogenesis in Arabidopsis thaliana. PLoS ONE 2013, 8, e69261. [Google Scholar] [CrossRef] [Green Version]
- Wickramasuriya, A.M.; Dunwell, J.M. Global scale transcriptome analysis of Arabidopsis embryogenesis in vitro. BMC Genom. 2015, 16, 301. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Y.C.; Zhang, T.Y.; Yu, Y.; Gou, L.P.; Yang, J.T.; Xu, J.; Pi, E.X. Regulatory Mechanisms of bHLH Transcription Factors in Plant Adaptive Responses to Various Abiotic Stresses. Front. Plant Sci. 2021, 12, 677611. [Google Scholar] [CrossRef]
- Nowak, K.; Wójcikowska, B.; Gaj, M.D. ERF022 impacts the induction of somatic embryogenesis in Arabidopsis through the ethylene-related pathway. Planta 2015, 241, 967–985. [Google Scholar] [CrossRef] [Green Version]
- Nowak, K.; Gaj, M.D. Stress-related function of bHLH109 in somatic embryo induction in Arabidopsis. J. Plant Physiol. 2016, 193, 119–126. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.H.; Zhao, X.Y.; Liu, Y.B.; Zhang, C.L.; O’Neill, S.D.; Zhang, X.S. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 2009, 59, 448–460. [Google Scholar] [CrossRef] [Green Version]
- Ayil-Gutiérrez, B.; Galaz-Ávalos, R.M.; Peña-Cabrera, E.; Loyola-Vargas, V.M. Dynamics of the concentration of IAA and some of its conjugates during the induction of somatic embryogenesis in Coffea canephora. Plant Signal. Behav. 2013, 8, e26998. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Wrobel-Marek, J.; Heidmann, I.; Horstman, A.; Chen, B.; Reis, R.; Angenent, G.C.; Boutilier, K. Auxin biosynthesis maintains embryo identity and growth during BABY BOOM-induced somatic embryogenesis. Plant Physiol. 2022, 188, 1095–1110. [Google Scholar] [CrossRef]
- Michalczuk, L.; Cooke, T.J.; Cohen, J.D. Auxin levels at different stages of carrot somatic embryogenesis. Phytochemistry 1992, 31, 1097–1103. [Google Scholar] [CrossRef]
- Chung, H.-H.; Chen, J.-T.; Chang, W.-C. Cytokinins induce direc somatic embryogenesis of Dendrobium chiengmai pink and subsequent plant regeneration. In Vitro Cell. Dev. Biol.-Plant 2005, 41, 765–769. [Google Scholar] [CrossRef]
- Kuklin, A. Ethylene Impact on Somatic Embryogenesis: Biotechnological Considerations. Biotechnol. Biotechnol. Equip. 1995, 9, 12–19. [Google Scholar] [CrossRef]
- Bell, L.; Trigiano, R.; Conger, B. Relationship of abscisic acid to somatic embryogenesis in Dactylis glomerata. Environ. Exp. Bot. 1993, 33, 495–499. [Google Scholar] [CrossRef]
- Elhiti, M.; Stasolla, C. Transduction of Signals during Somatic Embryogenesis. Plants 2022, 11, 178. [Google Scholar] [CrossRef]
- Camacho-Cristóbal, J.J.; Anzellotti, D.; González-Fontes, A. Changes in phenolic metabolism of tobacco plants during short-term boron deficiency. Plant Physiol. Biochem. 2002, 40, 997–1002. [Google Scholar] [CrossRef]
- Kong, G.-H.; Wu, Y.-P.; Shi, J.-L.; Xiang, N.-J.; Liu, L.-X.; Yang, G.-R.; Li, Y.-K.; Lu, X.-P.; Liu, Q.; Hu, Q.-F. Anti-tobacco mosaic virus phenylpropanoids from the stems of Nicotiana tabacum. Phytochem. Lett. 2015, 14, 230–233. [Google Scholar] [CrossRef]
- Fritz, C.; Palacios-Rojas, N.; Feil, R.; Stitt, M. Regulation of secondary metabolism by the carbon-nitrogen status in tobacco: Nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J. 2006, 46, 533–548. [Google Scholar] [CrossRef]
- Wu, H.; Qu, X.; Dong, Z.; Luo, L.; Shao, C.; Forner, J.; Lohmann, J.U.; Su, M.; Xu, M.; Liu, X.; et al. WUSCHEL triggers innate antiviral immunity in plant stem cells. Science 2020, 370, 227–231. [Google Scholar] [CrossRef]
- Zavattieri, M.A.; Frederico, A.M.; Lima, M.; Sabino, R.; Arnholdt-Schmitt, B. Induction of somatic embryogenesis as an example of stress-related plant reactions. Electron. J. Biotechnol. 2010, 13, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Juarez-Escobar, J.; Bojórquez-Velázquez, E.; Elizalde-Contreras, J.M.; Guerrero-Analco, J.A.; Loyola-Vargas, V.M.; Mata-Rosas, M.; Ruiz-May, E. Current Proteomic and Metabolomic Knowledge of Zygotic and Somatic Embryogenesis in Plants. Int. J. Mol. Sci. 2021, 22, 11807. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Henzler, C.; Hothorn, M. Molecular Mechanism for Plant Steroid Receptor Activation by Somatic Embryogenesis Co-Receptor Kinases. Science 2013, 341, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Nosaki, S.; Kaneko, M.K.; Tsuruta, F.; Yoshida, H.; Kato, Y.; Miura, K. Prevention of necrosis caused by transient expression in Nicotiana benthamiana by application of ascorbic acid. Plant Physiol. 2021, 186, 832–835. [Google Scholar] [CrossRef]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848–853. [Google Scholar] [CrossRef]
- Shinbo, Y.; Nakamura, Y.; Altaf-Ul-Amin, M.; Asahi, H.; Kurokawa, K.; Arita, M.; Saito, K.; Ohta, D.; Shibata, D.; Kanaya, S. KNApSAcK: A Comprehensive Species-Metabolite Relationship Database. In Plant Metabolomics; Saito, K., Dixon, R.A., Willmitzer, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 165–181. [Google Scholar]
- Zeng, X.; Zhang, P.; Wang, Y.; Qin, C.; Chen, S.; He, W.; Tao, L.; Tan, Y.; Gao, D.; Wang, B.; et al. CMAUP: A database of collective molecular activities of useful plants. Nucleic Acids Res. 2019, 47, D1118–D1127. [Google Scholar] [CrossRef] [Green Version]
- Feunang, Y.D.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Liang, S.; Luo, B.; Zhou, Z.; Bao, J.; Fang, R.; Wang, F.; Song, X.; Liao, Z.; Chen, G.; et al. Transcriptomic and Metabolomic Investigation on Leaf Necrosis Induced by ZmWus2 Transient Overexpression in Nicotiana benthamiana. Int. J. Mol. Sci. 2023, 24, 11190. https://doi.org/10.3390/ijms241311190
Zhang X, Liang S, Luo B, Zhou Z, Bao J, Fang R, Wang F, Song X, Liao Z, Chen G, et al. Transcriptomic and Metabolomic Investigation on Leaf Necrosis Induced by ZmWus2 Transient Overexpression in Nicotiana benthamiana. International Journal of Molecular Sciences. 2023; 24(13):11190. https://doi.org/10.3390/ijms241311190
Chicago/Turabian StyleZhang, Xianwen, Shuang Liang, Biao Luo, Zhongjing Zhou, Jiandong Bao, Ruiqiu Fang, Fang Wang, Xijiao Song, Zhenfeng Liao, Guang Chen, and et al. 2023. "Transcriptomic and Metabolomic Investigation on Leaf Necrosis Induced by ZmWus2 Transient Overexpression in Nicotiana benthamiana" International Journal of Molecular Sciences 24, no. 13: 11190. https://doi.org/10.3390/ijms241311190