Free Cholesterol Accelerates Aβ Self-Assembly on Membranes at Physiological Concentration
Abstract
:1. Introduction
2. Results
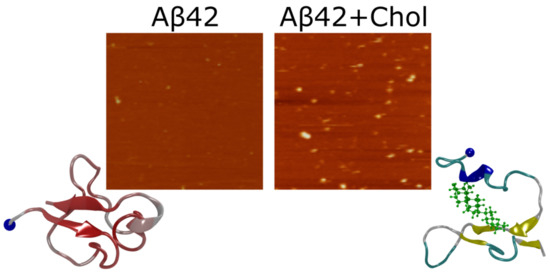
2.1. Rapid Appearance of Aggregates in Presence of Free Cholesterol
2.2. Acceleration of Aβ(1-42) Aggregation by Cholesterol inside Membrane
2.3. Dynamics of Aβ(1-42) Aggregation in Presence of Free Cholesterol
2.4. Computer Simulation of Interactions of Aβ(1-42) with Free Cholesterol
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of Supported Lipid Bilayer
4.3. Preparation of Aβ42 Protein Solution
4.4. Time-Lapse AFM Imaging
4.5. AFM Data Analysis
4.6. Molecular Dynamics Simulations
4.7. Analysis of MD Trajectories
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, S10–S17. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J. Has the Amyloid Cascade Hypothesis for Alzheimers Disease been Proved? Curr. Alzheimer Res. 2006, 3, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Crick, S.L.; Bu, G.; Frieden, C.; Pappu, R.V.; Lee, J.M. Amyloid seeds formed by cellular uptake, concentration, and aggregation of the amyloid-beta peptide. Proc. Natl. Acad. Sci. USA 2009, 106, 20324–20329. [Google Scholar] [CrossRef] [Green Version]
- Grimmer, T.; Riemenschneider, M.; Förstl, H.; Henriksen, G.; Klunk, W.E.; Mathis, C.A.; Shiga, T.; Wester, H.-J.; Kurz, A.; Drzezga, A. Beta Amyloid in Alzheimer’s Disease: Increased Deposition in Brain Is Reflected in Reduced Concentration in Cerebrospinal Fluid. Biol. Psychiatry 2009, 65, 927–934. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Hashemi, M.; Lv, Z.; Maity, S.; Rochet, J.C.; Lyubchenko, Y.L. A novel pathway for amyloids self-assembly in aggregates at nanomolar concentration mediated by the interaction with surfaces. Sci. Rep. 2017, 7, 45592. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Banerjee, S.; Zagorski, K.; Shlyakhtenko, L.S.; Kolomeisky, A.B.; Lyubchenko, Y.L. Molecular Model for the Surface-Catalyzed Protein Self-Assembly. J. Phys. Chem. B 2019, 124, 366–372. [Google Scholar] [CrossRef]
- Matsubara, T.; Nishihara, M.; Yasumori, H.; Nakai, M.; Yanagisawa, K.; Sato, T. Size and Shape of Amyloid Fibrils Induced by Ganglioside Nanoclusters: Role of Sialyl Oligosaccharide in Fibril Formation. Langmuir 2017, 33, 13874–13881. [Google Scholar] [CrossRef]
- Chi, E.Y.; Frey, S.L.; Lee, K.Y.C. Ganglioside GM1-Mediated Amyloid-beta Fibrillogenesis and Membrane Disruption. Biochemistry 2007, 46, 1913–1924. [Google Scholar] [CrossRef]
- Matsubara, T.; Yasumori, H.; Ito, K.; Shimoaka, T.; Hasegawa, T.; Sato, T. Amyloid-β fibrils assembled on ganglioside-enriched membranes contain both parallel β-sheets and turns. J. Biol. Chem. 2018, 293, 14146–14154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, S.; Hashemi, M.; Zagorski, K.; Lyubchenko, Y.L. Cholesterol in Membranes Facilitates Aggregation of Amyloid β Protein at Physiologically Relevant Concentrations. ACS Chem. Neurosci. 2021, 12, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Hicks, D.A.; Nalivaeva, N.N.; Turner, A.J. Lipid Rafts and Alzheimer’s Disease: Protein-Lipid Interactions and Perturbation of Signaling. Front. Physiol. 2012, 3, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahyayauch, H.; de la Arada, I.; Masserini, M.E.; Arrondo, J.L.R.; Goñi, F.M.; Alonso, A. The Binding of Aβ42 Peptide Monomers to Sphingomyelin/Cholesterol/Ganglioside Bilayers Assayed by Density Gradient Ultracentrifugation. Int. J. Mol. Sci. 2020, 21, 1674. [Google Scholar] [CrossRef] [Green Version]
- Fantini, J.; Yahi, N. Molecular insights into amyloid regulation by membrane cholesterol and sphingolipids: Common mechanisms in neurodegenerative diseases. Expert Rev. Mol. Med. 2010, 12, e27. [Google Scholar] [CrossRef]
- Popp, J.; Meichsner, S.; Kölsch, H.; Lewczuk, P.; Maier, W.; Kornhuber, J.; Jessen, F.; Lütjohann, D. Cerebral and extracerebral cholesterol metabolism and CSF markers of Alzheimer’s disease. Biochem. Pharmacol. 2013, 86, 37–42. [Google Scholar] [CrossRef]
- Panchal, M.; Loeper, J.; Cossec, J.-C.; Perruchini, C.; Lazar, A.; Pompon, D.; Duyckaerts, C. Enrichment of cholesterol in microdissected Alzheimer’s disease senile plaques as assessed by mass spectrometry. J. Lipid Res. 2010, 51, 598–605. [Google Scholar] [CrossRef] [Green Version]
- Gellermann, G.P.; Appel, T.R.; Davies, P.; Diekmann, S. Paired helical filaments contain small amounts of cholesterol, phosphatidylcholine and sphingolipids. Biol. Chem. 2006, 387, 1267–1274. [Google Scholar] [CrossRef]
- Ullrich, C.; Pirchl, M.; Humpel, C. Hypercholesterolemia in rats impairs the cholinergic system and leads to memory deficits. Mol. Cell. Neurosci. 2010, 45, 408–417. [Google Scholar] [CrossRef]
- Tansey, M.G.; Paul, R.; Choudhury, A.; Kumar, S.; Giri, A.; Sandhir, R.; Borah, A. Cholesterol contributes to dopamine-neuronal loss in MPTP mouse model of Parkinson’s disease: Involvement of mitochondrial dysfunctions and oxidative stress. PLoS ONE 2017, 12, e0171285. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, A.M.; Ferreira, A. Increased Membrane Cholesterol Might Render Mature Hippocampal Neurons More Susceptible to β-Amyloid-Induced Calpain Activation and Tau Toxicity. J. Neurosci. 2009, 29, 4640–4651. [Google Scholar] [CrossRef] [PubMed]
- Kiskis, J.; Fink, H.; Nyberg, L.; Thyr, J.; Li, J.-Y.; Enejder, A. Plaque-associated lipids in Alzheimer’s diseased brain tissue visualized by nonlinear microscopy. Sci. Rep. 2015, 5, 13489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, I.C.; Kuperstein, I.; Wilkinson, H.; Maes, E.; Vanbrabant, M.; Jonckheere, W.; Van Gelder, P.; Hartmann, D.; D’Hooge, R.; De Strooper, B.; et al. Lipids revert inert Aβ amyloid fibrils to neurotoxic protofibrils that affect learning in mice. EMBO 2007, 27, 224–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparr, E.; Linse, S. Lipid-protein interactions in amyloid formation. Biochim. Biophys. Acta 2019, 1867, 455–457. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Hashemi, M.; Zagorski, K.; Lyubchenko, Y.L. Interaction of Aβ42 with Membranes Triggers the Self-Assembly into Oligomers. Int. J. Mol. Sci. 2020, 21, 1129. [Google Scholar] [CrossRef] [Green Version]
- Lv, Z.; Banerjee, S.; Zagorski, K.; Lyubchenko, Y.L. Supported Lipid Bilayers for Atomic Force Microscopy Studies. Methods Mol. Biol. 2018, 1814, 129–143. [Google Scholar] [CrossRef]
- Lv, Z.; Hashemi, M.; Banerjee, S.; Zagorski, K.; Rochet, J.-C.; Lyubchenko, Y.L. Assembly of α-synuclein aggregates on phospholipid bilayers. Biochim. Biophys. Acta 2019, 1867, 802–812. [Google Scholar] [CrossRef]
- Di Scala, C.; Chahinian, H.; Yahi, N.; Garmy, N.; Fantini, J. Interaction of Alzheimer’s β-amyloid peptides with cholesterol: Mechanistic insights into amyloid pore formation. Biochemistry 2014, 53, 4489–4502. [Google Scholar] [CrossRef]
- Barrett Paul, J.; Song, Y.; Van Horn Wade, D.; Hustedt Eric, J.; Schafer Johanna, M.; Hadziselimovic, A.; Beel Andrew, J.; Sanders Charles, R. The Amyloid Precursor Protein Has a Flexible Transmembrane Domain and Binds Cholesterol. Science 2012, 336, 1168–1171. [Google Scholar] [CrossRef] [Green Version]
- Di Scala, C.; Yahi, N.; Lelièvre, C.; Garmy, N.; Chahinian, H.; Fantini, J. Biochemical Identification of a Linear Cholesterol-Binding Domain within Alzheimer’s β Amyloid Peptide. ACS Chem. Neurosci. 2013, 4, 509–517. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Zheng, J. Cholesterol Promotes the Interaction of Alzheimer β-Amyloid Monomer with Lipid Bilayer. J. Mol. Biol. 2012, 421, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Nguyen, P.H.; Derreumaux, P. Cholesterol Molecules Alter the Energy Landscape of Small Aβ1–42 Oligomers. J. Phys. Chem. B 2021, 125, 2299–2307. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Condron, M.M.; Teplow, D.B. Structure-neurotoxicity relationships of amyloid β-protein oligomers. Proc. Natl. Acad. Sci. USA 2009, 106, 14745–14750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fantini, J.; Di Scala, C.; Yahi, N.; Troadec, J.-D.; Sadelli, K.; Chahinian, H.; Garmy, N. Bexarotene Blocks Calcium-Permeable Ion Channels Formed by Neurotoxic Alzheimer’s β-Amyloid Peptides. ACS Chem. Neurosci. 2014, 5, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Cramer Paige, E.; Cirrito John, R.; Wesson Daniel, W.; Lee, C.Y.D.; Karlo, J.C.; Zinn Adriana, E.; Casali Brad, T.; Restivo Jessica, L.; Goebel Whitney, D.; James Michael, J.; et al. ApoE-Directed Therapeutics Rapidly Clear β-Amyloid and Reverse Deficits in AD Mouse Models. Science 2012, 335, 1503–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasumoto, T.; Takamura, Y.; Tsuji, M.; Watanabe-Nakayama, T.; Imamura, K.; Inoue, H.; Nakamura, S.; Inoue, T.; Kimura, A.; Yano, S.; et al. High molecular weight amyloid β 1-42 oligomers induce neurotoxicity via plasma membrane damage. FASEB J. 2019, 33, 9220–9234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treiber-Held, S.; Distl, R.; Meske, V.; Albert, F.; Ohm, T.G. Spatial and temporal distribution of intracellular free cholesterol in brains of a Niemann–Pick type C mouse model showing hyperphosphorylated tau protein. Implications for Alzheimer’s disease. J. Pathol. 2003, 200, 95–103. [Google Scholar] [CrossRef]
- Zhang, Y.; Hashemi, M.; Lv, Z.; Lyubchenko, Y.L. Self-assembly of the full-length amyloid Abeta42 protein in dimers. Nanoscale 2016, 8, 18928–18937. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [Green Version]
- Dickson, C.J.; Madej, B.D.; Skjevik, Å.A.; Betz, R.M.; Teigen, K.; Gould, I.R.; Walker, R.C. Lipid14: The Amber Lipid Force Field. J. Chem. Theory Comput. 2014, 10, 865–879. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cisneros, G.A.; Cruzeiro, V.W.D.; Darden, T.A.; et al. Amber 1; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cisneros, G.A.; Cruzeiro, V.W.D.; Darden, T.A.; et al. Amber 20; University of California: San Francisco, CA, USA, 2020. [Google Scholar]
- Glykos, N.M. Software news and updates carma: A molecular dynamics analysis program. J. Comput. Chem. 2006, 27, 1765–1768. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashemi, M.; Banerjee, S.; Lyubchenko, Y.L. Free Cholesterol Accelerates Aβ Self-Assembly on Membranes at Physiological Concentration. Int. J. Mol. Sci. 2022, 23, 2803. https://doi.org/10.3390/ijms23052803
Hashemi M, Banerjee S, Lyubchenko YL. Free Cholesterol Accelerates Aβ Self-Assembly on Membranes at Physiological Concentration. International Journal of Molecular Sciences. 2022; 23(5):2803. https://doi.org/10.3390/ijms23052803
Chicago/Turabian StyleHashemi, Mohtadin, Siddhartha Banerjee, and Yuri L. Lyubchenko. 2022. "Free Cholesterol Accelerates Aβ Self-Assembly on Membranes at Physiological Concentration" International Journal of Molecular Sciences 23, no. 5: 2803. https://doi.org/10.3390/ijms23052803