Dynamics of Endothelial Engagement and Filopodia Formation in Complex 3D Microscaffolds
Abstract
:1. Introduction
2. Results
2.1. Fabrication of Hexagonal Microstructures and Cell Seeding

2.2. Formation of an Endothelial Top Monolayer on Top of the Structures, and its Orientation in the Function of the Geometry of the Underlying Microstructure
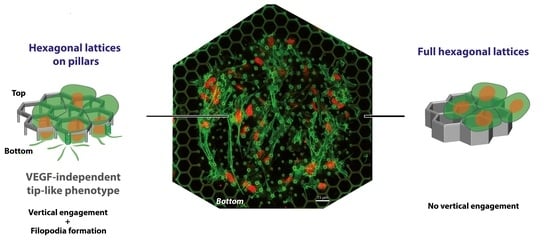
2.3. Endothelial Engagement from the Top Monolayer in the “Open” Microstructures Only
2.3.1. Global Observations on Endothelial Cell Engagement
2.3.2. Quantification of Nuclei Engagement in Function of the Microtopography
2.4. Induction of Endothelial Filopodia in Open Microstructures: Development of Automated Detection and Tracking, and Filopodia Characteristics
2.5. Molecular Mechanisms Governing the Transitions between the Different Protrusive Modes Characteristic for the Microstructures
2.5.1. Reversible Switches between Mesenchymal (Filopodia + Dactylopodia-Like) and Amoeboid Modes Are Observed, with ROCK and MLCK Activation of Actomyosin Contractility Favouring Distinct Migration Modes
- Spontaneous mesenchymal–bleb transitions
- Mesenchymal–bleb reversible transitions in function of ROCK/MLCK balance
2.5.2. Dactylopodia-Like Protrusions and Their Stabilization by FAK Inhibition
2.6. Independence on VEGF
3. Discussion
3.1. Summary
3.2. Vertical Engagement Inside the Microstructures
3.3. Confinement and the Amoeboid Protrusive Mode
3.4. Filopodia Generation and Induction of Dactylopodia-Like Protrusions
3.5. Signalling Pathways
3.6. Perspectives
4. Materials and Methods
4.1. Two-Photon Polymerization
4.2. Cell Culture and Treatments
4.3. Cell Labelling
4.4. Imaging and Image Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Development of Algorithms for Nuclei Quantification on Fixed Samples

Appendix A.2. Development of Algorithms for Filopodia Detection, Tracking and Quantifications
Appendix A.2.1. Filopodia Detection on Fixed Samples

Appendix A.2.2. CNN-Based Automated Detection and Tracking on Filopodia in Live Experiments
- ▪
- Constitution of a training dataset
- ▪
- CNN model
- ▪
- Segmentation and tracking
Appendix A.3. Additional Information for Nuclei Quantification
Appendix A.4. Early Colonization, and Membrane Labelling

Appendix A.5. Filopodia Orientation in Elongated Structures

Appendix A.6. Filopodia Lengths and Orientations in Regular Structures

Appendix A.7. Additional Information for Filopodia Quantification
Appendix A.8. Additional Data on ML-7, Y-27632, PF-573228 and Sunitinib Effects


Appendix A.9. VEGF Accessibility

References
- Bettinger, C.J.; Langer, R.; Borenstein, J.T. Engineering Substrate Topography at the Micro- and Nanoscale to Control Cell Function. Angew. Chem. Int. Ed. 2009, 48, 5406–5415. [Google Scholar] [CrossRef] [Green Version]
- Karina Kulangaraa, K.W.L. Substrate topography shapes cell function. Soft Matter. 2009, 5, 4072–4076. [Google Scholar] [CrossRef]
- Flemming, R.; Murphy, C.; Abrams, G.; Goodman, S.; Nealey, P. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials 1999, 20, 573–588. [Google Scholar] [CrossRef]
- Teixeira, A.; Abrams, G.A.; Bertics, P.J.; Murphy, C.J.; Nealey, P.F. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J. Cell Sci. 2003, 116, 1881–1892. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Jones, J.A.; Xu, Y.; Low, H.-Y.; Anderson, J.M.; Leong, K.W. Characterization of topographical effects on macrophage behavior in a foreign body response model. Biomaterials 2010, 31, 3479–3491. [Google Scholar] [CrossRef] [PubMed]
- Webb, P.; Clark, P.; Skepper, J.; Compston, A.; Wood, A. Guidance of oligodendrocytes and their progenitors by substratum topography. J. Cell Sci. 1995, 108 Part 8, 2747–2760. [Google Scholar] [CrossRef]
- Wood, A. Contact guidance on microfabricated substrata: The response of teleost fin mesenchyme cells to repeating topographical patterns. J. Cell Sci. 1988, 90, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Rajnicek, A.; Britland, S.; McCaig, C. Contact guidance of CNS neurites on grooved quartz: Influence of groove dimensions, neuronal age and cell type. J. Cell Sci. 1997, 110 Part 2, 2905–2913. [Google Scholar] [CrossRef]
- Gomez, N.; Lu, Y.; Chen, S.; Schmidt, C.E. Immobilized nerve growth factor and microtopography have distinct effects on polarization versus axon elongation in hippocampal cells in culture. Biomaterials 2007, 28, 271–284. [Google Scholar] [CrossRef]
- Hanson, J.N.; Motala, M.J.; Heien, M.L.; Gillette, M.; Sweedler, J.; Nuzzo, R.G. Textural guidance cues for controlling process outgrowth of mammalian neurons. Lab Chip 2009, 9, 122–131. [Google Scholar] [CrossRef] [Green Version]
- Dalton, B.A.; Walboomers, X.F.; Dziegielewski, M.; Evans, M.D.; Taylor, S.; Jansen, J.A.; Steele, J.G. Modulation of epithelial tissue and cell migration by microgrooves. J. Biomed. Mater. Res. 2001, 56, 195–207. [Google Scholar] [CrossRef]
- Bettinger, C.J.; Zhang, Z.; Gerecht, S.; Borenstein, J.T.; Langer, R. Enhancement of In Vitro Capillary Tube Formation by Substrate Nanotopography. Adv. Mater. 2007, 20, 99–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greiner, A.M.; Jäckel, M.; Scheiwe, A.C.; Stamow, D.R.; Autenrieth, T.J.; Lahann, J.; Franz, C.M.; Bastmeyer, M. Multifunctional polymer scaffolds with adjustable pore size and chemoattractant gradients for studying cell matrix invasion. Biomaterials 2014, 35, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.-C.; Chiu, K.-H. Inverted colloidal crystal scaffolds with laminin-derived peptides for neuronal differentiation of bone marrow stromal cells. Biomaterials 2011, 32, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Koroleva, A.; Deiwick, A.; Nguyen, A.; Schlie-Wolter, S.; Narayan, R.; Timashev, P.; Popov, V.; Bagratashvili, V.N.; Chichkov, B. Osteogenic Differentiation of Human Mesenchymal Stem Cells in 3-D Zr-Si Organic-Inorganic Scaffolds Produced by Two-Photon Polymerization Technique. PLoS ONE 2015, 10, e0118164. [Google Scholar] [CrossRef]
- Yim, E.; Pang, S.; Leong, K.W. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp. Cell Res. 2007, 313, 1820–1829. [Google Scholar] [CrossRef] [Green Version]
- Mata, A.; Kim, E.J.; Boehm, C.A.; Fleischman, A.J.; Muschler, G.F.; Roy, S. A three-dimensional scaffold with precise micro-architecture and surface micro-textures. Biomaterials 2009, 30, 4610–4617. [Google Scholar] [CrossRef] [Green Version]
- Grewal, M.G.; Highley, C.B. Electrospun hydrogels for dynamic culture systems: Advantages, progress, and opportunities. Biomater. Sci. 2021, 9, 4228–4245. [Google Scholar] [CrossRef]
- Dolega, M.E.; Wagh, J.; Gerbaud, S.; Kermarrec, F.; Alcaraz, J.-P.; Martin, D.K.; Gidrol, X.; Picollet-D’hahan, N. Facile Bench-Top Fabrication of Enclosed Circular Microchannels Provides 3D Confined Structure for Growth of Prostate Epithelial Cells. PLoS ONE 2014, 9, e99416. [Google Scholar] [CrossRef]
- Venzac, B.; Madoun, R.; Benarab, T.; Monnier, S.; Cayrac, F.; Myram, S.; Leconte, L.; Amblard, F.; Viovy, J.-L.; Descroix, S.; et al. Engineering small tubes with changes in diameter for the study of kidney cell organization. Biomicrofluidics 2018, 12, 24114. [Google Scholar] [CrossRef]
- Homan, K.A.; Kolesky, D.B.; Skylar-Scott, M.A.; Herrmann, J.; Obuobi, H.; Moisan, A.; Lewis, J.A. Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips. Sci. Rep. 2016, 6, 34845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.S.; Kim, R.H.; Yang, D.Y.; Park, S.H. Advances in 3D nano/microfabrication using two-photon initiated photopolymerization. Prog. Polym. Sci. 2008, 33, 631–681. [Google Scholar] [CrossRef]
- Gittard, S.D.; Narayan, R.J. Laser direct writing of micro- and nano-scale medical devices. Expert Rev. Med Devices 2010, 7, 343–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hippler, M.; Lemma, E.D.; Bertels, S.; Blasco, E.; Barner-Kowollik, C.; Wegener, M.; Bastmeyer, M. 3D Scaffolds to Study Basic Cell Biology. Adv. Mater. 2019, 31, e1808110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spagnolo, B.; Brunetti, V.; Leménager, G.; De Luca, E.; Sileo, L.; Pellegrino, T.; Pompa, P.P.; De Vittorio, M.; Pisanello, F. Three-dimensional cage-like microscaffolds for cell invasion studies. Sci. Rep. 2015, 5, srep10531. [Google Scholar] [CrossRef] [Green Version]
- Scheiwe, A.C.; Frank, S.C.; Autenrieth, T.J.; Bastmeyer, M.; Wegener, M. Subcellular stretch-induced cytoskeletal response of single fibroblasts within 3D designer scaffolds. Biomaterials 2015, 44, 186–194. [Google Scholar] [CrossRef]
- Sprague, E.A.; Tio, F.; Ahmed, S.H.; Granada, J.F.; Bailey, S.R. Impact of Parallel Micro-Engineered Stent Grooves on Endothelial Cell Migration, Proliferation, and Function. Circ. Cardiovasc. Interv. 2012, 5, 499–507. [Google Scholar] [CrossRef] [Green Version]
- Bedair, T.M.; ElNaggar, M.A.; Joung, Y.K.; Han, D.K. Recent advances to accelerate re-endothelialization for vascular stents. J. Tissue Eng. 2017, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Lutter, C.; Nothhaft, M.; Rzany, A.; Garlichs, C.D.; Cicha, I. Effect of specific surface microstructures on substrate endothelialisation and thrombogenicity: Importance for stent design. Clin. Hemorheol. Microcirc. 2015, 59, 219–233. [Google Scholar] [CrossRef]
- Deveza, L.; Choi, J.; Yang, F. Therapeutic Angiogenesis for Treating Cardiovascular Diseases. Theranostics 2012, 2, 801–814. [Google Scholar] [CrossRef]
- Gorin, C.; Rochefort, G.Y.; Bascetin, R.; Ying, H.; Lesieur, J.; Sadoine, J.; Beckouche, N.; Berndt, S.; Novais, A.; Lesage, M.; et al. Priming Dental Pulp Stem Cells with Fibroblast Growth Factor-2 Increases Angiogenesis of Implanted Tissue-Engineered Constructs Through Hepatocyte Growth Factor and Vascular Endothelial Growth Factor Secretion. STEM CELLS Transl. Med. 2016, 5, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Tiemeijer, L.A.; Frimat, J.-P.; Stassen, O.; Bouten, C.; Sahlgren, C.M. Spatial patterning of the Notch ligand Dll4 controls endothelial sprouting in vitro. Sci. Rep. 2018, 8, 6392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, S.; Lin, S.; Cheung, C.; Yim, E.K.; Toh, Y.-C. Topography elicits distinct phenotypes and functions in human primary and stem cell derived endothelial cells. Biomaterials 2020, 234, 119747. [Google Scholar] [CrossRef] [PubMed]
- Mogilner, A.; Rubinstein, B. The Physics of Filopodial Protrusion. Biophys. J. 2005, 89, 782–795. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, A.M.; Barbacena, P.; Russo, A.; Vaccaro, S.; Ramalho, D.; Pena, A.; Lima, A.P.; Ferreira, R.R.; Fidalgo, M.A.; El-Marjou, F.; et al. Endothelial cell invasion is controlled by dactylopodia. Proc. Natl. Acad. Sci. USA 2021, 118, e2023829118. [Google Scholar] [CrossRef] [PubMed]
- Germain, S.; Monnot, C.; Muller, L.; Eichmann, A. Hypoxia-driven angiogenesis: Role of tip cells and extracellular matrix scaffolding. Curr. Opin. Hematol. 2010, 17, 245–251. [Google Scholar] [CrossRef]
- Coscoy, S.; Baïz, S.; Octon, J.; Rhoné, B.; Perquis, L.; Tseng, Q.; Amblard, F.; Semetey, V. Microtopographies control the development of basal protrusions in epithelial sheets. Biointerphases 2018, 13, 41003. [Google Scholar] [CrossRef]
- Bray, M.-A.P.; Adams, W.J.; Geisse, N.A.; Feinberg, A.W.; Sheehy, S.P.; Parker, K.K. Nuclear morphology and deformation in engineered cardiac myocytes and tissues. Biomaterials 2010, 31, 5143–5150. [Google Scholar] [CrossRef] [Green Version]
- Caille, N.; Tardy, Y.; Meister, J.J. Assessment of Strain Field in Endothelial Cells Subjected to Uniaxial Deformation of Their Substrate. Ann. Biomed. Eng. 1998, 26, 409–416. [Google Scholar] [CrossRef]
- Maniotis, A.J.; Chen, C.; Ingber, D.E. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA 1997, 94, 849–854. [Google Scholar] [CrossRef] [Green Version]
- Angulo-Urarte, A.; van der Wal, T.; Huveneers, S. Cell-cell junctions as sensors and transducers of mechanical forces. Biochim. Biophys. Acta (BBA)—Biomembr. 2020, 1862, 183316. [Google Scholar] [CrossRef] [PubMed]
- Cazes, A.; Galaup, A.; Chomel, C.; Bignon, M.; Bréchot, N.; Le Jan, S.; Weber, H.; Corvol, P.; Muller, L.; Germain, S.; et al. Extracellular Matrix–Bound Angiopoietin-Like 4 Inhibits Endothelial Cell Adhesion, Migration, and Sprouting and Alters Actin Cytoskeleton. Circ. Res. 2006, 99, 1207–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbančič, V.; Butler, R.; Richier, B.; Peter, M.; Mason, J.; Livesey, F.J.; Holt, C.E.; Gallop, J.L. Filopodyan: An open-source pipeline for the analysis of filopodia. J. Cell Biol. 2017, 216, 3405–3422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacquemet, G.; Paatero, I.; Carisey, A.F.; Padzik, A.; Orange, J.S.; Hamidi, H.; Ivaska, J. FiloQuant reveals increased filopodia density during DCIS progression. bioRxiv 2017. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Uchida, Y.; Wei, T.; Liu, C.; Adams, R.H.; Kubota, Y.; Gutkind, J.S.; Mukouyama, Y.-S.; Adelstein, R.S. Nonmuscle myosin 2 regulates cortical stability during sprouting angiogenesis. Mol. Biol. Cell 2020, 31, 1974–1987. [Google Scholar] [CrossRef]
- Lomakin, A.J.; Cattin, C.J.; Cuvelier, D.; Alraies, Z.; Molina, M.; Nader, G.P.F.; Srivastava, N.; Sáez, P.J.; Garcia-Arcos, J.M.; Zhitnyak, I.Y.; et al. The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science 2020, 370, eaba2894. [Google Scholar] [CrossRef]
- Ghosh, I.; Singh, R.K.; Mishra, M.; Kapoor, S.; Jana, S.S. Switching between blebbing and lamellipodia depends on the degree of non-muscle myosin II activity. J. Cell Sci. 2021, 134, jcs248732. [Google Scholar] [CrossRef]
- Totsukawa, G.; Wu, Y.; Sasaki, Y.; Hartshorne, D.J.; Yamakita, Y.; Yamashiro, S.; Matsumura, F. Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J. Cell Biol. 2004, 164, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Wehrle-Haller, B.; Vogel, V. Maturation of Filopodia Shaft Adhesions Is Upregulated by Local Cycles of Lamellipodia Advancements and Retractions. PLoS ONE 2014, 9, e107097. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Gong, J.; Zhen, Y. Focal adhesion kinase is activated by microtubule-depolymerizing agents and regulates membrane blebbing in human endothelial cells. J. Cell. Mol. Med. 2020, 24, 7228–7238. [Google Scholar] [CrossRef]
- Hopkins, A.M.; Pineda, A.A.; Winfree, L.M.; Brown, G.T.; Laukoetter, M.G.; Nusrat, A. Organized migration of epithelial cells requires control of adhesion and protrusion through Rho kinase effectors. Am. J. Physiol. Liver Physiol. 2007, 292, G806–G817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, J.T.; Morgan, P.; Dowell, N.; Leu, B. Myosin light chain phosphorylation and growth cone motility. J. Neurobiol. 2002, 52, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, M.; Sainson, R.C.A.; Pérez-Del-Pulgar, S.; Aoto, J.N.; Aitkenhead, M.; Taylor, K.L.; Carpenter, P.M.; Hughes, C.C.W. VEGF121 and VEGF165 Regulate Blood Vessel Diameter Through Vascular Endothelial Growth Factor Receptor 2 in an in vitro Angiogenesis Model. Lab. Investig. 2003, 83, 1873–1885. [Google Scholar] [CrossRef]
- Beckouche, N.; Bignon, M.; Lelarge, V.; Mathivet, T.; Pichol-Thievend, C.; Berndt, S.; Hardouin, J.; Garand, M.; Ardidie-Robouant, C.; Barret, A.; et al. The interaction of heparan sulfate proteoglycans with endothelial transglutaminase-2 limits VEGF 165 -induced angiogenesis. Sci. Signal. 2015, 8, ra70. [Google Scholar] [CrossRef]
- Labots, M.; Gotink, K.J.; Dekker, H.; Azijli, K.; Van Der Mijn, J.C.; Huijts, C.M.; Piersma, S.R.; Jiménez, C.R.; Verheul, H.M.W. Evaluation of a tyrosine kinase peptide microarray for tyrosine kinase inhibitor therapy selection in cancer. Exp. Mol. Med. 2016, 48, e279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Mijn, J.C.; Broxterman, H.J.; Knol, J.C.; Piersma, S.R.; De Haas, R.R.; Dekker, H.; Pham, T.V.; Van Beusechem, V.W.; Halmos, B.; Mier, J.W.; et al. Sunitinib activates Axl signaling in renal cell cancer. Int. J. Cancer 2016, 138, 3002–3010. [Google Scholar] [CrossRef] [Green Version]
- Guetta-Terrier, C.; Monzo, P.; Zhu, J.; Long, H.; Venkatraman, L.; Zhou, Y.; Wang, P.; Chew, S.Y.; Mogilner, A.; Ladoux, B.; et al. Protrusive waves guide 3D cell migration along nanofibers. J. Cell Biol. 2015, 211, 683–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, N.; Bertulli, C.; Sadok, A.; Huang, Y.Y.S. Dynamics of filopodium-like protrusion and endothelial cellular motility on one-dimensional extracellular matrix fibrils. Interface Focus 2014, 4, 20130060. [Google Scholar] [CrossRef] [Green Version]
- Doyle, A.D.; Wang, F.W.; Matsumoto, K.; Yamada, K.M. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 2009, 184, 481–490. [Google Scholar] [CrossRef]
- Yevick, H.G.; Duclos, G.; Bonnet, I.; Silberzan, P. Architecture and migration of an epithelium on a cylindrical wire. Proc. Natl. Acad. Sci. USA 2015, 112, 5944–5949. [Google Scholar] [CrossRef] [Green Version]
- Herrera, A.; Herrera, M.; Guerra-Perez, N.; Galindo-Pumariño, C.; Larriba, M.J.; García-Barberán, V.; Gil, B.; Giménez-Moyano, S.; Ferreiro-Monteagudo, R.; Veguillas, P.; et al. Endothelial cell activation on 3D-matrices derived from PDGF-BB-stimulated fibroblasts is mediated by Snail1. Oncogenesis 2018, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Charles-Orszag, A.; Tsai, F.-C.; Bonazzi, D.; Manriquez, V.; Sachse, M.; Mallet, A.; Salles, A.; Melican, K.; Staneva, R.; Bertin, A.; et al. Adhesion to nanofibers drives cell membrane remodeling through one-dimensional wetting. Nat. Commun 2018, 9, 4450–4463. [Google Scholar] [CrossRef]
- Du, X.; Wang, Y.; Yuan, L.; Weng, Y.; Chen, G.; Hu, Z. Guiding the behaviors of human umbilical vein endothelial cells with patterned silk fibroin films. Colloids Surf. B Biointerfaces 2014, 122, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Formentín, P.; Catalán, Ú.; Pol, L.; Fernández-Castillejo, S.; Solà, R.; Marsal, L.F. Collagen and fibronectin surface modification of nanoporous anodic alumina and macroporous silicon for endothelial cell cultures. J. Biol. Eng. 2018, 12, 21. [Google Scholar] [CrossRef]
- Beltrán-Partida, E.; Valdéz-Salas, B.; Moreno-Ulloa, A.; Escamilla, A.; Curiel, M.A.; Rosales-Ibáñez, R.; Villarreal, F.; Bastidas, D.M.; Bastidas, J.M. Improved in vitro angiogenic behavior on anodized titanium dioxide nanotubes. J. Nanobiotechnol. 2017, 15, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albuschies, J.; Vogel, V. The role of filopodia in the recognition of nanotopographies. Sci. Rep. 2013, 3, srep01658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhuri, P.; Harfouche, R.; Soni, S.; Hentschel, D.M.; Sengupta, S. Shape Effect of Carbon Nanovectors on Angiogenesis. ACS Nano 2010, 4, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Borghi, N.; Alias, K.; De Gennes, P.-G.; Brochard-Wyart, F. Wetting fibers with liposomes. J. Colloid Interface Sci. 2005, 285, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, X.; Wu, J.; Wong, T.M.; Feng, X.; Yang, C.; Wu, S.; Zheng, Y.; Liu, X.; Cheung, K.M.C.; et al. Micro- and Nanohemispherical 3D Imprints Modulate the Osteogenic Differentiation and Mineralization Tendency of Bone Cells. ACS Appl. Mater. Interfaces 2019, 11, 35513–35524. [Google Scholar] [CrossRef]
- Li, J.-R.; Shi, L.; Deng, Z.; Lo, S.H.; Liu, G.-Y. Nanostructures of Designed Geometry and Functionality Enable Regulation of Cellular Signaling Processes. Biochemistry 2012, 51, 5876–5893. [Google Scholar] [CrossRef]
- Nune, K.C.; Misra, R.; Gai, X.; Li, S.J.; Hao, Y.L. Surface nanotopography-induced favorable modulation of bioactivity and osteoconductive potential of anodized 3D printed Ti-6Al-4V alloy mesh structure. J. Biomater. Appl. 2017, 32, 1032–1048. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Luo, D.; Liu, Y. Effect of the nano/microscale structure of biomaterial scaffolds on bone regeneration. Int. J. Oral Sci. 2020, 12, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phng, L.-K.; Stanchi, F.; Gerhardt, H. Filopodia are dispensable for endothelial tip cell guidance. Development 2013, 140, 4031–4040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fantin, A.; Lampropoulou, A.; Gestri, G.; Raimondi, C.; Senatore, V.; Zachary, I.; Ruhrberg, C. NRP1 Regulates CDC42 Activation to Promote Filopodia Formation in Endothelial Tip Cells. Cell Rep. 2015, 11, 1577–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamalice, L.; Houle, F.; Jourdan, G.; Huot, J. Phosphorylation of tyrosine 1214 on VEGFR2 is required for VEGF-induced activation of Cdc42 upstream of SAPK2/p38. Oncogene 2004, 23, 434–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, C.G.; Moroishi, T.; Guan, K.-L. YAP and TAZ: A nexus for Hippo signaling and beyond. Trends Cell Biol. 2015, 25, 499–513. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Wang, K.; Gu, X.; Leong, K.W. Biophysical Regulation of Cell Behavior—Cross Talk between Substrate Stiffness and Nanotopography. Engineering 2017, 3, 36–54. [Google Scholar] [CrossRef]
- Kim, M.-C.; Silberberg, Y.R.; Abeyaratne, R.; Kamm, R.D.; Asada, H.H. Computational modeling of three-dimensional ECM-rigidity sensing to guide directed cell migration. Proc. Natl. Acad. Sci. USA 2018, 115, E390–E399. [Google Scholar] [CrossRef] [Green Version]
- Atlas, Y.; Gorin, C.; Novais, A.; Marchand, M.F.; Chatzopoulou, E.; Lesieur, J.; Bascetin, R.; Binet-Moussy, C.; Sadoine, J.; Lesage, M.; et al. Microvascular maturation by mesenchymal stem cells in vitro improves blood perfusion in implanted tissue constructs. Biomaterials 2020, 268, 120594. [Google Scholar] [CrossRef]
- Wang, I.; Bouriau, M.; Baldeck, P.; Martineau, C.; Andraud, C. Three-dimensional microfabrication by two-photon-initiated polymerization with a low-cost microlaser. Opt. Lett. 2002, 27, 1348–1350. [Google Scholar] [CrossRef]
- Kervrann, C.; Boulanger, J. Optimal Spatial Adaptation for Patch-Based Image Denoising. IEEE Trans. Image Process. 2006, 15, 2866–2878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, B.; Schindelin, J.; Cardona, A.; Longair, M.; Heisenberg, M. A high-level 3D visualization API for Java and ImageJ. BMC Bioinform. 2010, 11, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Ehling, M.; März, S.; Seebach, J.; Tarbashevich, K.; Sixta, T.; Pitulescu, M.E.; Werner, A.-C.; Flach, B.; Montanez, E.; et al. Polarized actin and VE-cadherin dynamics regulate junctional remodelling and cell migration during sprouting angiogenesis. Nat. Commun. 2017, 8, 2210. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| Quantity | Extremity | Value |
|---|---|---|
| Length (μm) | ||
| 3.36 ± 0.41 | ||
| Lifetime (s) | ||
| 233 ± 58 | ||
| Elongation speed (nm/s) | (+)-end | 45.8 ± 5.8 |
| (−)-end | 44.3 ± 5.4 | |
| Retraction speed (nm/s) | (+)-end (−)-end | 48.6 ± 7.0 |
| 42.4 ± 5.0 | ||
| Normalized number (μm−1) | 0.037 ± 0.020 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ucla, P.; Ju, X.; Demircioglu, M.; Baiz, S.; Muller, L.; Germain, S.; Monnot, C.; Semetey, V.; Coscoy, S. Dynamics of Endothelial Engagement and Filopodia Formation in Complex 3D Microscaffolds. Int. J. Mol. Sci. 2022, 23, 2415. https://doi.org/10.3390/ijms23052415
Ucla P, Ju X, Demircioglu M, Baiz S, Muller L, Germain S, Monnot C, Semetey V, Coscoy S. Dynamics of Endothelial Engagement and Filopodia Formation in Complex 3D Microscaffolds. International Journal of Molecular Sciences. 2022; 23(5):2415. https://doi.org/10.3390/ijms23052415
Chicago/Turabian StyleUcla, Pierre, Xingming Ju, Melisa Demircioglu, Sarah Baiz, Laurent Muller, Stéphane Germain, Catherine Monnot, Vincent Semetey, and Sylvie Coscoy. 2022. "Dynamics of Endothelial Engagement and Filopodia Formation in Complex 3D Microscaffolds" International Journal of Molecular Sciences 23, no. 5: 2415. https://doi.org/10.3390/ijms23052415