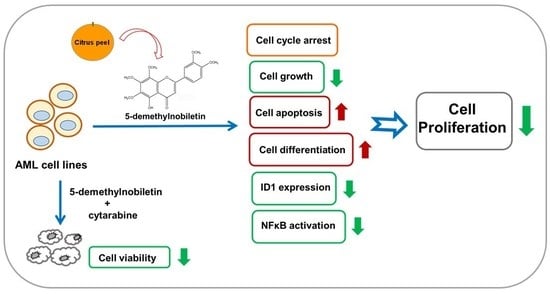
5-Demethylnobiletin Inhibits Cell Proliferation, Downregulates ID1 Expression, Modulates the NF-κB/TNF-α Pathway and Exerts Antileukemic Effects in AML Cells
Abstract
:1. Introduction
2. Results
2.1. Effects of 5-Demethyl NOB on Human Leukemia Cell Viability
2.2. Effects of 5-Demethyl NOB on Cell Cycle Progression in THP-1 Cells
2.3. 5-Demethyl NOB Induced Cell Apoptosis and Differentiation in AML Cell Lines
2.4. Analysis of 5-Demethyl NOB-Modulated Gene Expression in AML Cells
2.5. Analysis of 5-Demethyl NOB-Regulated Biological Processes (BPs)
2.6. Analysis of Gene Set Enrichment in 5-Demethyl NOB-Treated THP-1 Cells
2.7. Effects of Combined Treatment with 5-Demethyl NOB and Cytarabine in AML Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Analysis of Cell Viability
4.4. Flow Cytometric Analysis of Cell Cycle Progression and Apoptotic Cells
4.5. Western Blot Analysis
4.6. Reverse-Transcription Quantitative Polymerase Chain Reaction (RT–qPCR) Analysis
4.7. RNA Preparation and cDNA Microarray Analysis
4.8. Gene Ontology and Gene Set Enrichment Analysis (GSEA)
4.9. Preparation of ID1 Promoter-Reporter Constructs, Plasmid Transfection, and Luciferase Reporter Assay
4.10. Combination Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dohner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, E.M.; Tallman, M.S. Emerging therapeutic drugs for AML. Blood 2016, 127, 71–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Rooij, J.D.; Zwaan, C.M.; van den Heuvel-Eibrink, M. Pediatric AML: From Biology to Clinical Management. J. Clin. Med. 2015, 4, 127–149. [Google Scholar] [CrossRef] [PubMed]
- Dohner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [Green Version]
- McMahon, C.M.; Ferng, T.; Canaani, J.; Wang, E.S.; Morrissette, J.J.D.; Eastburn, D.J.; Pellegrino, M.; Durruthy-Durruthy, R.; Watt, C.D.; Asthana, S.; et al. Clonal Selection with RAS Pathway Activation Mediates Secondary Clinical Resistance to Selective FLT3 Inhibition in Acute Myeloid Leukemia. Cancer Discov. 2019, 9, 1050–1063. [Google Scholar] [CrossRef]
- Quek, L.; David, M.D.; Kennedy, A.; Metzner, M.; Amatangelo, M.; Shih, A.; Stoilova, B.; Quivoron, C.; Heiblig, M.; Willekens, C.; et al. Clonal heterogeneity of acute myeloid leukemia treated with the IDH2 inhibitor enasidenib. Nat. Med. 2018, 24, 1167–1177. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; Kadia, T.M.; DiNardo, C.D.; Welch, M.A.; Ravandi, F. Acute myeloid leukemia: Treatment and research outlook for 2021 and the MD Anderson approach. Cancer 2021, 127, 1186–1207. [Google Scholar] [CrossRef]
- Carter, J.L.; Hege, K.; Yang, J.; Kalpage, H.A.; Su, Y.; Edwards, H.; Huttemann, M.; Taub, J.W.; Ge, Y. Targeting multiple signaling pathways: The new approach to acute myeloid leukemia therapy. Signal Transduct. Target. Ther. 2020, 5, 288. [Google Scholar] [CrossRef]
- Binder, S.; Luciano, M.; Horejs-Hoeck, J. The cytokine network in acute myeloid leukemia (AML): A focus on pro- and anti-inflammatory mediators. Cytokine Growth Factor Rev. 2018, 43, 8–15. [Google Scholar] [CrossRef]
- Fisher, D.A.C.; Fowles, J.S.; Zhou, A.; Oh, S.T. Inflammatory Pathophysiology as a Contributor to Myeloproliferative Neoplasms. Front. Immunol. 2021, 12, 683401. [Google Scholar] [CrossRef]
- Hwang, D.; Kim, M.; Park, H.; Jeong, M.I.; Jung, W.; Kim, B. Natural Products and Acute Myeloid Leukemia: A Review Highlighting Mechanisms of Action. Nutrients 2019, 11, 1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Wu, Q.; Zhou, F. Targeting acute myeloid leukemia stem cells: Current therapies in development and potential strategies with new dimensions. Crit. Rev. Oncol. Hematol. 2020, 152, 102993. [Google Scholar] [CrossRef]
- Cotoraci, C.; Ciceu, A.; Sasu, A.; Miutescu, E.; Hermenean, A. The Anti-Leukemic Activity of Natural Compounds. Molecules 2021, 26, 2709. [Google Scholar] [CrossRef] [PubMed]
- Koolaji, N.; Shammugasamy, B.; Schindeler, A.; Dong, Q.; Dehghani, F.; Valtchev, P. Citrus Peel Flavonoids as Potential Cancer Prevention Agents. Curr. Dev. Nutr. 2020, 4, nzaa025. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Chen, Y.T.; Gao, W.Y.; Wu, M.J.; Yen, J.H. Nobiletin Down-Regulates c-KIT Gene Expression and Exerts Antileukemic Effects on Human Acute Myeloid Leukemia Cells. J. Agric. Food Chem. 2018, 66, 13423–13434. [Google Scholar] [CrossRef]
- Yen, J.H.; Lin, C.Y.; Chuang, C.H.; Chin, H.K.; Wu, M.J.; Chen, P.Y. Nobiletin Promotes Megakaryocytic Differentiation through the MAPK/ERK-Dependent EGR1 Expression and Exerts Anti-Leukemic Effects in Human Chronic Myeloid Leukemia (CML) K562 Cells. Cells 2020, 9, 877. [Google Scholar] [CrossRef] [Green Version]
- Goh, J.X.H.; Tan, L.T.; Goh, J.K.; Chan, K.G.; Pusparajah, P.; Lee, L.H.; Goh, B.H. Nobiletin and Derivatives: Functional Compounds from Citrus Fruit Peel for Colon Cancer Chemoprevention. Cancers 2019, 11, 867. [Google Scholar] [CrossRef] [Green Version]
- Qiu, P.; Dong, P.; Guan, H.; Li, S.; Ho, C.T.; Pan, M.H.; McClements, D.J.; Xiao, H. Inhibitory effects of 5-hydroxy polymethoxyflavones on colon cancer cells. Mol. Nutr. Food Res. 2010, 54 (Suppl. S2), S244–S252. [Google Scholar] [CrossRef]
- Tung, Y.C.; Li, S.; Huang, Q.; Hung, W.L.; Ho, C.T.; Wei, G.J.; Pan, M.H. 5-Demethylnobiletin and 5-Acetoxy-6,7,8,3′,4′-pentamethoxyflavone Suppress Lipid Accumulation by Activating the LKB1-AMPK Pathway in 3T3-L1 Preadipocytes and High Fat Diet-Fed C57BL/6 Mice. J. Agric. Food Chem. 2016, 64, 3196–3205. [Google Scholar] [CrossRef]
- Wang, M.; Meng, D.; Zhang, P.; Wang, X.; Du, G.; Brennan, C.; Li, S.; Ho, C.T.; Zhao, H. Antioxidant Protection of Nobiletin, 5-Demethylnobiletin, Tangeretin, and 5-Demethyltangeretin from Citrus Peel in Saccharomyces cerevisiae. J. Agric. Food Chem. 2018, 66, 3155–3160. [Google Scholar] [CrossRef]
- Chiu, S.P.; Wu, M.J.; Chen, P.Y.; Ho, Y.R.; Tai, M.H.; Ho, C.T.; Yen, J.H. Neurotrophic action of 5-hydroxylated polymethoxyflavones: 5-demethylnobiletin and gardenin A stimulate neuritogenesis in PC12 cells. J. Agric. Food Chem. 2013, 61, 9453–9463. [Google Scholar] [CrossRef] [PubMed]
- Yen, J.H.; Weng, C.Y.; Li, S.; Lo, Y.H.; Pan, M.H.; Fu, S.H.; Ho, C.T.; Wu, M.J. Citrus flavonoid 5-demethylnobiletin suppresses scavenger receptor expression in THP-1 cells and alters lipid homeostasis in HepG2 liver cells. Mol. Nutr. Food Res. 2011, 55, 733–748. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.M.; Qu, L.Q.; Ng, J.P.L.; Zeng, W.; Yu, L.; Song, L.L.; Wong, V.K.W.; Xia, C.L.; Law, B.Y.K. Natural Citrus flavanone 5-demethylnobiletin stimulates melanogenesis through the activation of cAMP/CREB pathway in B16F10 cells. Phytomed. Int. J. Phytother. Phytopharm. 2022, 98, 153941. [Google Scholar] [CrossRef]
- Yang, G.; Li, S.; Yang, Y.; Yuan, L.; Wang, P.; Zhao, H.; Ho, C.T.; Lin, C.C. Nobiletin and 5-Hydroxy-6,7,8,3′,4′-pentamethoxyflavone Ameliorate 12-O-Tetradecanoylphorbol-13-acetate-Induced Psoriasis-Like Mouse Skin Lesions by Regulating the Expression of Ki-67 and Proliferating Cell Nuclear Antigen and the Differentiation of CD4(+) T Cells through Mitogen-Activated Protein Kinase Signaling Pathways. J. Agric. Food Chem. 2018, 66, 8299–8306. [Google Scholar] [PubMed]
- Chou, Y.C.; Lin, Y.H.; Lin, P.H.; Tung, Y.C.; Ho, C.T.; Pan, M.H. Dietary 5-demethylnobiletin modulates xenobiotic-metabolizing enzymes and ameliorates colon carcinogenesis in benzo[a]pyrene-induced mice. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2021, 155, 112380. [Google Scholar] [CrossRef]
- Song, M.; Lan, Y.; Wu, X.; Han, Y.; Wang, M.; Zheng, J.; Li, Z.; Li, F.; Zhou, J.; Xiao, J.; et al. The chemopreventive effect of 5-demethylnobiletin, a unique citrus flavonoid, on colitis-driven colorectal carcinogenesis in mice is associated with its colonic metabolites. Food Funct. 2020, 11, 4940–4952. [Google Scholar] [CrossRef]
- Song, M.; Charoensinphon, N.; Wu, X.; Zheng, J.; Gao, Z.; Xu, F.; Wang, M.; Xiao, H. Inhibitory Effects of Metabolites of 5-Demethylnobiletin on Human Nonsmall Cell Lung Cancer Cells. J. Agric. Food Chem. 2016, 64, 4943–4949. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Zhang, H.; Chen, J.; Cao, J.; Chen, Q.; Li, X.; Sun, C. Polymethoxyflavones from citrus inhibited gastric cancer cell proliferation through inducing apoptosis by upregulating RARbeta, both in vitro and in vivo. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2020, 146, 111811. [Google Scholar] [CrossRef]
- Li, S.; Pan, M.H.; Lai, C.S.; Lo, C.Y.; Dushenkov, S.; Ho, C.T. Isolation and syntheses of polymethoxyflavones and hydroxylated polymethoxyflavones as inhibitors of HL-60 cell lines. Bioorg. Med. Chem. 2007, 15, 3381–3389. [Google Scholar] [CrossRef]
- Lai, C.S.; Wu, J.C.; Ho, C.T.; Pan, M.H. Disease chemopreventive effects and molecular mechanisms of hydroxylated polymethoxyflavones. BioFactors 2015, 41, 301–313. [Google Scholar] [CrossRef]
- Joshi, A.D. New Insights Into Physiological and Pathophysiological Functions of Stanniocalcin 2. Front. Endocrinol. (Lausanne) 2020, 11, 172. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Lui, H.M.; Lin, S.L.; Lee, J.M.; Ying, S.Y. Regulation of cell proliferation, apoptosis, and carcinogenesis by activin. Exp. Biol. Med. 2002, 227, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Aleman-Muench, G.R.; Soldevila, G. When versatility matters: Activins/inhibins as key regulators of immunity. Immunol. Cell Biol. 2012, 90, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Ling, M.T.; Wang, X.; Zhang, X.; Wong, Y.C. The multiple roles of Id-1 in cancer progression. Differ. Res. Biol. Divers. 2006, 74, 481–487. [Google Scholar] [CrossRef]
- Perk, J.; Iavarone, A.; Benezra, R. Id family of helix-loop-helix proteins in cancer. Nat. Rev. Cancer 2005, 5, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Bo, Z.; Gong, W.; Guo, Y. Inhibitor of Differentiation 1 (Id1) in Cancer and Cancer Therapy. Int. J. Med. Sci. 2020, 17, 995–1005. [Google Scholar] [CrossRef] [Green Version]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Zambon, A.C.; Gaj, S.; Ho, I.; Hanspers, K.; Vranizan, K.; Evelo, C.T.; Conklin, B.R.; Pico, A.R.; Salomonis, N. GO-Elite: A flexible solution for pathway and ontology over-representation. Bioinformatics 2012, 28, 2209–2210. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.S.; Wang, C.Y.; Chen, P.S.; Hung, J.H.; Yen, J.H.; Wu, M.J. 8-Hydroxydaidzein Downregulates JAK/STAT, MMP, Oxidative Phosphorylation, and PI3K/AKT Pathways in K562 Cells. Biomedicines 2021, 9, 1907. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Z.; Zhou, J. Tumor necrosis factor alpha in the onset and progression of leukemia. Exp. Hematol. 2017, 45, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Kagoya, Y.; Yoshimi, A.; Kataoka, K.; Nakagawa, M.; Kumano, K.; Arai, S.; Kobayashi, H.; Saito, T.; Iwakura, Y.; Kurokawa, M. Positive feedback between NF-kappaB and TNF-alpha promotes leukemia-initiating cell capacity. J. Clin. Investig. 2014, 124, 528–542. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.K.; Wang, H.C.; Ho, C.T.; Chen, H.Y.; Li, S.; Chan, H.L.; Chung, T.W.; Tan, K.T.; Li, Y.R.; Lin, C.C. 5-demethylnobiletin promotes the formation of polymerized tubulin, leads to G2/M phase arrest and induces autophagy via JNK activation in human lung cancer cells. J. Nutr. Biochem. 2015, 26, 484–504. [Google Scholar] [CrossRef]
- Song, M.; Wu, X.; Charoensinphon, N.; Wang, M.; Zheng, J.; Gao, Z.; Xu, F.; Li, Z.; Li, F.; Zhou, J.; et al. Dietary 5-demethylnobiletin inhibits cigarette carcinogen NNK-induced lung tumorigenesis in mice. Food Funct. 2017, 8, 954–963. [Google Scholar] [CrossRef]
- Donjerkovic, D.; Scott, D.W. Regulation of the G1 phase of the mammalian cell cycle. Cell Res. 2000, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.T.; Li, S.; Li, Y.R.; Cheng, S.L.; Lin, S.H.; Tung, Y.T. Synergistic Anticancer Effect of a Combination of Paclitaxel and 5-Demethylnobiletin Against Lung Cancer Cell Line In Vitro and In Vivo. Appl. Biochem. Biotechnol. 2019, 187, 1328–1343. [Google Scholar] [CrossRef]
- Akao, Y.; Itoh, T.; Ohguchi, K.; Iinuma, M.; Nozawa, Y. Interactive effects of polymethoxy flavones from Citrus on cell growth inhibition in human neuroblastoma SH-SY5Y cells. Bioorg. Med. Chem. 2008, 16, 2803–2810. [Google Scholar] [CrossRef]
- Zhang, M.; Pan, Y.; Dong, Q.; Tang, X.; Xin, Y.; Yin, B.; Zhu, J.; Kou, X.; Ho, C.T.; Huang, Q. Development of organogel-based emulsions to enhance the loading and bioaccessibility of 5-demethylnobiletin. Food Res. Int. 2021, 148, 110592. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, Y.; Wu, Z.; Zhang, L.; He, D.; Li, X.; Wang, Z. Synergistic combination therapy of lung cancer: Cetuximab functionalized nanostructured lipid carriers for the co-delivery of paclitaxel and 5-Demethylnobiletin. Biomed. Pharmacother. Biomed. Pharmacother. 2019, 118, 109225. [Google Scholar] [CrossRef]
- Yao, M.; Li, Z.; Julian McClements, D.; Tang, Z.; Xiao, H. Design of nanoemulsion-based delivery systems to enhance intestinal lymphatic transport of lipophilic food bioactives: Influence of oil type. Food Chem. 2020, 317, 126229. [Google Scholar] [CrossRef]
- Cao, X.; Ma, C.; Gao, Z.; Zheng, J.; He, L.; McClements, D.J.; Xiao, H. Characterization of the Interactions between Titanium Dioxide Nanoparticles and Polymethoxyflavones Using Surface-Enhanced Raman Spectroscopy. J. Agric. Food Chem. 2016, 64, 9436–9441. [Google Scholar] [CrossRef] [PubMed]
- Roschger, C.; Cabrele, C. The Id-protein family in developmental and cancer-associated pathways. Cell Commun. Signal. 2017, 15, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, F.; Kang, B.; Sun, X.H. Id proteins: Small molecules, mighty regulators. Curr. Top. Dev. Biol. 2014, 110, 189–216. [Google Scholar]
- Fong, S.; Debs, R.J.; Desprez, P.Y. Id genes and proteins as promising targets in cancer therapy. Trends Mol. Med. 2004, 10, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Hirsch, P.; Fava, F.; Lapusan, S.; Marzac, C.; Teyssandier, I.; Pardo, J.; Marie, J.P.; Legrand, O. High Id1 expression is associated with poor prognosis in 237 patients with acute myeloid leukemia. Blood 2009, 114, 2993–3000. [Google Scholar] [CrossRef] [Green Version]
- Man, N.; Sun, X.J.; Tan, Y.; Garcia-Cao, M.; Liu, F.; Cheng, G.; Hatlen, M.; Xu, H.; Shah, R.; Chastain, N.; et al. Differential role of Id1 in MLL-AF9-driven leukemia based on cell of origin. Blood 2016, 127, 2322–2326. [Google Scholar] [CrossRef] [Green Version]
- Qian, Y.; Chen, X. ID1, inhibitor of differentiation/DNA binding, is an effector of the p53-dependent DNA damage response pathway. J. Biol. Chem. 2008, 283, 22410–22416. [Google Scholar] [CrossRef] [Green Version]
- Hemmati, S.; Haque, T.; Gritsman, K. Inflammatory Signaling Pathways in Preleukemic and Leukemic Stem Cells. Front. Oncol. 2017, 7, 265. [Google Scholar] [CrossRef] [Green Version]
- Guzman, M.L.; Rossi, R.M.; Karnischky, L.; Li, X.; Peterson, D.R.; Howard, D.S.; Jordan, C.T. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood 2005, 105, 4163–4169. [Google Scholar] [CrossRef]
- Darwish, N.H.E.; Sudha, T.; Godugu, K.; Bharali, D.J.; Elbaz, O.; El-Ghaffar, H.A.A.; Azmy, E.; Anber, N.; Mousa, S.A. Novel Targeted Nano-Parthenolide Molecule against NF-kB in Acute Myeloid Leukemia. Molecules 2019, 24, 2103. [Google Scholar] [CrossRef] [Green Version]
- Fajardo-Orduna, G.R.; Ledesma-Martinez, E.; Aguiniga-Sanchez, I.; Mora-Garcia, M.L.; Weiss-Steider, B.; Santiago-Osorio, E. Inhibitors of Chemoresistance Pathways in Combination with Ara-C to Overcome Multidrug Resistance in AML. A Mini Review. Int. J. Mol. Sci. 2021, 22, 4955. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sang, S.; Pan, M.H.; Lai, C.S.; Lo, C.Y.; Yang, C.S.; Ho, C.T. Anti-inflammatory property of the urinary metabolites of nobiletin in mouse. Bioorg. Med. Chem. Lett. 2007, 17, 5177–5181. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Chao, T.Y.; Hsu, H.J.; Wang, C.Y.; Lin, C.Y.; Gao, W.Y.; Wu, M.J.; Yen, J.H. The Lipid-Modulating Effect of Tangeretin on the Inhibition of Angiopoietin-like 3 (ANGPTL3) Gene Expression through Regulation of LXRalpha Activation in Hepatic Cells. Int. J. Mol. Sci. 2021, 22, 9853. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.S.; Yen, J.H.; Wang, C.Y.; Chen, P.Y.; Hung, J.H.; Wu, M.J. 8-Hydroxydaidzein, an Isoflavone from Fermented Soybean, Induces Autophagy, Apoptosis, Differentiation, and Degradation of Oncoprotein BCR-ABL in K562 Cells. Biomedicines 2020, 8, 506. [Google Scholar] [CrossRef]
- Nehlin, J.O.; Hara, E.; Kuo, W.L.; Collins, C.; Campisi, J. Genomic organization, sequence, and chromosomal localization of the human helix-loop-helix Id1 gene. Biochem. Biophys. Res. Commun. 1997, 231, 628–634. [Google Scholar] [CrossRef] [Green Version]
- Chou, T.C.; Talalay, P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzym. Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]













| Differentially Expressed Genes (DEGs) | 5-Demethyl NOB Treatment 48 h versus 0 h |
|---|---|
| Genes in upregulated expression | 552 |
| Genes in downregulated expression | 694 |
| Genes | Primers |
|---|---|
| CD11b | 5′-ACTTGCAGTGAGAACACGTATG-3′ 5′-AGAGCCATCAATCAAGAAGGC-3′ |
| INHBE | 5′-GGTCTGTGTGTCCCTCCTGT-3′ 5′-GGAGCTGTAGGCTGAAGTGG-3′ |
| ID1 | 5′-CGGATCGAGGGAGAACAAG-3′ 5′-TCCCACCCCCTAAAGTCTCT-3′ |
| GAPDH | 5′-CATGAGAAGTATGACAACAGCCT-3′ 5′-AGTCCTTCCACGATACCAAAGT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, P.-Y.; Wang, C.-Y.; Tsao, E.-C.; Chen, Y.-T.; Wu, M.-J.; Ho, C.-T.; Yen, J.-H. 5-Demethylnobiletin Inhibits Cell Proliferation, Downregulates ID1 Expression, Modulates the NF-κB/TNF-α Pathway and Exerts Antileukemic Effects in AML Cells. Int. J. Mol. Sci. 2022, 23, 7392. https://doi.org/10.3390/ijms23137392
Chen P-Y, Wang C-Y, Tsao E-C, Chen Y-T, Wu M-J, Ho C-T, Yen J-H. 5-Demethylnobiletin Inhibits Cell Proliferation, Downregulates ID1 Expression, Modulates the NF-κB/TNF-α Pathway and Exerts Antileukemic Effects in AML Cells. International Journal of Molecular Sciences. 2022; 23(13):7392. https://doi.org/10.3390/ijms23137392
Chicago/Turabian StyleChen, Pei-Yi, Chih-Yang Wang, En-Ci Tsao, Yu-Ting Chen, Ming-Jiuan Wu, Chi-Tang Ho, and Jui-Hung Yen. 2022. "5-Demethylnobiletin Inhibits Cell Proliferation, Downregulates ID1 Expression, Modulates the NF-κB/TNF-α Pathway and Exerts Antileukemic Effects in AML Cells" International Journal of Molecular Sciences 23, no. 13: 7392. https://doi.org/10.3390/ijms23137392