Regulation of Plant Responses to Salt Stress
Abstract
:1. Introduction
2. Salt Stress Sensing
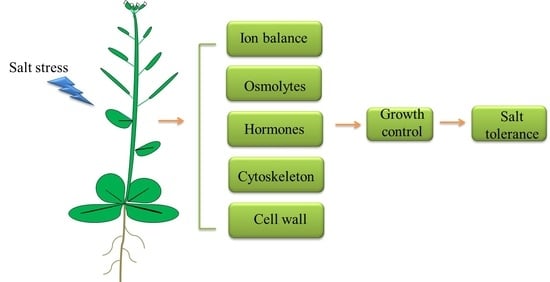
3. Regulation of Plant Response to Salt Stress
3.1. Ion Balance
3.2. Osmotic Homeostasis
3.3. Phytohormone Signaling Mediation—ABA Signaling and BR Signaling
3.4. Cytoskeleton Functions
3.5. Cell Wall Regulation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Savary, S.; Akter, S.; Almekinders, C.; Harris, J.; Korsten, L.; Rötter, R.; Waddington, S.; Watson, D. Mapping disruption and resilience mechanisms in food systems. Food Secur. 2020, 12, 695–717. [Google Scholar] [CrossRef]
- Hazell, P.B.R.; Wood, S. Drivers of change in global agriculture. Philos. Trans. R. Soc. B Biol. Sci. 2007, 363, 495–515. [Google Scholar] [CrossRef] [Green Version]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, W.-Y.; Yun, A.D.-J. A New Insight of Salt Stress Signaling in Plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef]
- Ziska, L.H.; Bunce, J.A.; Shimono, H.; Gealy, D.R.; Baker, J.T.; Newton, P.C.D.; Reynolds, M.P.; Jagadish, K.S.V.; Zhu, C.; Howden, M.; et al. Food security and climate change: On the potential to adapt global crop production by active selection to rising atmospheric carbon dioxide. Proc. R. Soc. B Boil. Sci. 2012, 279, 4097–4105. [Google Scholar] [CrossRef]
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An Intracellular Degradation Pathway Regulating Plant Survival and Stress Response. Front. Plant Sci. 2020, 11, 164. [Google Scholar] [CrossRef] [Green Version]
- Horie, T.; Karahara, I.; Katsuhara, M. Salinity tolerance mechanisms in glycophytes: An overview with the central focus on rice plants. Rice 2012, 5, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a Putative Calcium Sensor, Interacts with the Protein Kinase SOS2 to Protect Arabidopsis Shoots from Salt Stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.J.; Kim, W.-Y.; Yun, D.-J. A role for GIGANTEA. Plant Signal. Behav. 2013, 8, e24820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, Z. Plant abiotic stress: New insights into the factors that activate and modulate plant responses. J. Integr. Plant Biol. 2021, 63, 429–430. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Hoehenwarter, W. Changes in the Phosphosproteome and Metabolome Link Early Signaling Events to Rearrangement of Photosynthesis and Central Metabolism in Salinity and Oxidative Stress Response in Arabidopsis. Plant Physiol. 2015, 69, 3021–3033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shumilina, J.; Kusnetsova, A.; Tsarev, A.; Van Rensburg, H.C.J.; Medvedev, S.; Demidchik, V.; Ende, W.V.D.; Frolov, A. Glycation of Plant Proteins: Regulatory Roles and Interplay with Sugar Signalling? Int. J. Mol. Sci. 2019, 20, 2366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nat. Cell Biol. 2014, 514, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wu, Q.-R.; Liu, L.-L.; Zhang, H.-M.; Gao, J.-W.; Pei, Z.-M. Osmotic stress alters circadian cytosolic Ca2+ oscillations and OSCA1 is required in circadian gated stress adaptation. Plant Signal. Behav. 2020, 15, 1836883. [Google Scholar] [CrossRef]
- Stephan, A.B.; Kunz, H.H.; Yang, E.; Schroeder, J.I. Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and invovlement of plastidial KEA transporters. Proc. Natl. Acad. Sci. USA 2016, 113, E5242–E5249. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nat. Cell Biol. 2019, 572, 341–346. [Google Scholar] [CrossRef]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.-C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I.; et al. The FERONIA Receptor Kinase Maintains Cell-Wall Integrity during Salt Stress through Ca2+ Signaling. Curr. Biol. 2018, 28, 666–675.e5. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Chai, X.; Gao, Q.; Zhou, L.; Zhang, S.; Li, L.; Luan, S. Dynamic Interactions of Plant CNGC Subunits and Calmodulins Drive Oscillatory Ca2+ Channel Activities. Dev. Cell 2019, 48, 710–725.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nat. Cell Biol. 2019, 572, 131–135. [Google Scholar] [CrossRef]
- Wu, F.; Chi, Y.; Jiang, Z.; Xu, Y.; Xie, L.; Huang, F.; Wan, D.; Ni, J.; Yuan, F.; Wu, X.; et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nat. Cell Biol. 2020, 578, 577–581. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Lin, H.; Chen, S.; Becker, K.; Yang, Y.; Zhao, J.; Kudla, J.; Schumaker, K.S.; Guo, Y. Inhibition of the Arabidopsis Salt Overly Sensitive Pathway by 14-3-3 Proteins. Plant Cell 2014, 26, 1166–1182. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.-Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.-Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1352. [Google Scholar] [CrossRef] [Green Version]
- Tan, T.; Cai, J.; Zhan, E.; Yang, Y.; Zhao, J.; Guo, Y.; Zhou, H. Stability and localization of 14-3-3 proteins are involved in salt tolerance in Arabidopsis. Plant Mol. Biol. 2016, 92, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Barajas-Lopez, J.D.D.; Moreno, J.R.; Gamez-Arjona, F.M.; Pardo, J.M.; Punkkinen, M.; Zhu, J.-K.; Quintero, F.J.; Fujii, H. Upstream kinases of plant SnRKs are involved in salt stress tolerance. Plant J. 2017, 93, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 Complex Generates and Fine-Tunes an AtANN4-Dependent Calcium Signature under Salt Stress. Dev. Cell 2019, 48, 697–709.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhou, H.; Zhang, Y.; Li, Z.; Yang, Y.; Guo, Y. The GSK3-like Kinase BIN2 Is a Molecular Switch between the Salt Stress Response and Growth Recovery in Arabidopsis thaliana. Dev. Cell 2020, 55, 367–380.e6. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.-L.; Qi, G.-N.; Feng, H.-Q.; Zhao, S.; Zhao, S.-S.; Wang, Y.; Wu, W.-H. Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+homeostasis in Arabidopsis. Plant J. 2013, 74, 258–266. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Alemán, F.; Martínez, V.; Rubio, F. K+ uptake in plant roots. The systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 2014, 171, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, H.-D.; Chen, L.-Q.; Wang, Y.; Liu, L.-L.; He, L.; Wu, W.-H. A Protein Kinase, Interacting with Two Calcineurin B-like Proteins, Regulates K+ Transporter AKT1 in Arabidopsis. Cell 2006, 125, 1347–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Barrena, M.J.; Chaves-Sanjuan, A.; Raddatz, N.; Mendoza, I.; Cortés, Á.; Gago, F.; González-Rubio, J.M.; Benavente, J.L.; Quintero, F.J.; Pardo, J.M.; et al. Recognition and Activation of the Plant AKT1 Potassium Channel by the Kinase CIPK23. Plant Physiol. 2020, 182, 2143–2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheong, Y.H.; Pandey, G.K.; Grant, J.J.; Batistic, O.; Li, L.; Kim, B.-G.; Lee, S.-C.; Kudla, J.; Luan, S. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007, 52, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Lan, W.-Z.; Kim, B.-G.; Li, L.; Cheong, Y.H.; Pandey, G.K.; Lu, G.; Buchanan, B.B.; Luan, S. A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. USA 2007, 104, 15959–15964. [Google Scholar] [CrossRef] [Green Version]
- Lan, W.Z.; Lee, S.C.; Che, Y.; Jiang, Y.; Luan, S. Mechanistic Analysis of AKT1 Regulation by the CBL–CIPK–PP2CA Interactions. Mol. Plant 2011, 4, 527–536. [Google Scholar] [CrossRef]
- Latz, A.; Becker, D.; Hekman, M.; Müller, T.; Beyhl, D.; Marten, I.; Eing, C.; Fischer, A.; Dunkel, M.; Bertl, A.; et al. TPK1, a Ca2+-regulated Arabidopsis vacuole two-pore K+ channel is activated by 14-3-3 proteins. Plant J. 2007, 52, 449–459. [Google Scholar] [CrossRef]
- Latz, A.; Mehlmer, N.; Zapf, S.; Mueller, T.D.; Wurzinger, B.; Pfister, B.; Csaszar, E.; Hedrich, R.; Teige, M.; Becker, D. Salt Stress Triggers Phosphorylation of the Arabidopsis Vacuolar K+ Channel TPK1 by Calcium-Dependent Protein Kinases (CDPKs). Mol. Plant 2013, 6, 1274–1289. [Google Scholar] [CrossRef] [Green Version]
- Ali, A.; Maggio, A.; Bressan, R.A.; Yun, D.-J. Role and Functional Differences of HKT1-Type Transporters in Plants under Salt Stress. Int. J. Mol. Sci. 2019, 20, 1059. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2017, 217, 523–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.-Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+exclusion and salt tolerance in maize. New Phytol. 2017, 217, 1161–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, M.; Chen, P.; Meng, S.; Xu, P.; Lan, W. The Arabidopsis phosphatase PP2C49 negatively regulates salt tolerance through inhibition of AtHKT1;1. J. Integr. Plant Biol. 2021, 63, 528–542. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Bülbül, S.; Piao, W.; Choi, G.; Paek, N. Arabidopsis EARLY FLOWERING 3 increases salt tolerance by suppressing salt stress response pathways. Plant J. 2017, 92, 1106–1120. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Wang, X.; He, Y.; Xu, H.; Wang, L. Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2;1 -mediated sodium homeostasis. EMBO J. 2021, 40, e105086. [Google Scholar] [CrossRef]
- Ayadi, M.; Brini, F.; Masmoudi, K. Overexpression of a Wheat Aquaporin Gene, TdPIP2;1, Enhances Salt and Drought Tolerance in Transgenic Durum Wheat cv. Maali. Int. J. Mol. Sci. 2019, 20, 2389. [Google Scholar] [CrossRef] [Green Version]
- Marusig, D.; Tombesi, S. Abscisic Acid Mediates Drought and Salt Stress Responses in Vitis vinifera—A Review. Int. J. Mol. Sci. 2020, 21, 8648. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef]
- Zhou, X.; Naguro, I.; Ichijo, H.; Watanabe, K. Mitogen-activated protein kinases as key players in osmotic stress signaling. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 2037–2052. [Google Scholar] [CrossRef]
- Moustafa, K.; AbuQamar, S.; Jarrar, M.; Al-Rajab, A.J.; Trémouillaux-Guiller, J. MAPK cascades and major abiotic stresses. Plant Cell Rep. 2014, 33, 1217–1225. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Zhang, M.; Kong, X.; Xing, X.; Liu, Y.; Zhou, Y.; Liu, Y.; Sun, L.; Li, D. ZmMPK17, a novel maize group D MAP kinase gene, is involved in multiple stress responses. Planta 2011, 235, 661–676. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, S.; Pan, J.; Kong, X.; Zhou, Y.; Liu, Y.; Li, D. The overexpression of a maize mitogen-activated protein kinase gene (ZmMPK5) confers salt stress tolerance and induces defence responses in tobacco. Plant Biol. 2013, 16, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, G.; Zhao, J.; Zhang, L.; Ai, L.; Han, Y.; Sun, D.; Zhang, S.; Sun, Y. The Receptor-Like Kinase SIT1 Mediates Salt Sensitivity by Activating MAPK3/6 and Regulating Ethylene Homeostasis in Rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Gao, J.; Sun, S.; Zhang, Z.; Yu, B.; Li, J.; Xie, C.; Li, G.; Wang, P.; Bressan, R.A.; et al. The Calcium-Responsive Phospholipid-Binding BONZAI Proteins Control Global Osmotic Stress Responses in Plants Through Repression of Immune Signaling. SSRN Electron. J. 2020, 30, 4815. [Google Scholar] [CrossRef]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2016, 90, 856–867. [Google Scholar] [CrossRef]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate Peroxidase and Catalase Activities and Their Genetic Regulation in Plants Subjected to Drought and Salinity Stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Liu, D.; Liu, S. Two rice cytosolic ascorbate peroxidases differentially improve salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2007, 26, 1909–1917. [Google Scholar] [CrossRef]
- Sharma, R.; Sahoo, A.; Devendran, R.; Jain, M. Over-Expression of a Rice Tau Class Glutathione S-Transferase Gene Improves Tolerance to Salinity and Oxidative Stresses in Arabidopsis. PLoS ONE 2014, 9, e92900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, N.; Chu, Y.; Chen, H.; Li, X.; Wu, Q.; Jin, L.; Wang, G.; Huang, J. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA–mediated regulatory pathway and ROS scavenging. PLoS Genet. 2018, 14, e1007662. [Google Scholar] [CrossRef] [Green Version]
- Seo, P.J.; Park, J.-M.; Kang, S.K.; Kim, S.-G.; Park, C.-M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 2010, 233, 189–200. [Google Scholar] [CrossRef]
- Chapman, J.M.; Muhlemann, J.K.; Gayomba, S.R.; Muday, G.K. RBOH-Dependent ROS Synthesis and ROS Scavenging by Plant Specialized Metabolites To Modulate Plant Development and Stress Responses. Chem. Res. Toxicol. 2019, 32, 370–396. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Wang, Y.; Zhang, S.; Zhang, J. Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J. Exp. Bot. 2002, 53, 2201–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L.; et al. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019, 287, 110188. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nat. Cell Biol. 2018, 556, 235–238. [Google Scholar] [CrossRef]
- Barrero, J.M.; Rodriguez, P.L.; Quesada, V.; Piqueras, P.; Ponce, M.R.; Micol, J.L. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 2006, 29, 2000–2008. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.-J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018, 23, 3340–3351.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.-C.; Liu, X.; Fu, L.; Hou, Y.-J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 2018, 69, 100–112.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Li, Y.; Zhang, Z.; Liu, X.; Hsu, C.; Du, Y.; Sang, T.; Zhu, C.; Wang, Y.; Satheesh, V.; et al. A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 2020, 11, 613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywińska, E.; Bucholc, M.; Kulik, A.; Ciesielski, A.; Lichocka, M.; Dębski, J.; Ludwików, A.; Dadlez, M.; Rodriguez, P.L.; Dobrowolska, G. Phosphatase ABI1 and okadaic acid-sensitive phosphoprotein phosphatases inhibit salt stress-activated SnRK2.4 kinase. BMC Plant Biol. 2016, 16, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Ohta, M.; Guo, Y.; Halfter, U.; Zhu, J.-K. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 2003, 100, 11771–11776. [Google Scholar] [CrossRef] [Green Version]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [Green Version]
- Su, Q.; Zheng, X.; Tian, Y.; Wang, C. Exogenous Brassinolide Alleviates Salt Stress in Malus hupehensis Rehd. by Regulating the Transcription of NHX-Type Na+(K+)/H+ Antiporters. Front. Plant Sci. 2020, 11, 38. [Google Scholar] [CrossRef]
- Liu, J.; Gao, H.; Zheng, Q.; Wang, C.; Wang, X.; Wang, Q. Effects of 24-epibrassinolide on plant growth, osmotic regulation and ion homeostasis of salt-stressed canola. Plant Biol. 2013, 16, 440–450. [Google Scholar] [CrossRef]
- Krishna, P.; Prasad, B.D.; Rahman, T. Brassinosteroid Action in Plant Abiotic Stress Tolerance. Methods Mol. Biol. 2017, 1564, 193–202. [Google Scholar] [CrossRef]
- Fàbregas, N.; Lozano-Elena, F.; Blasco-Escámez, D.; Tohge, T.; Martínez-Andújar, C.; Albacete, A.; Osorio, S.; Bustamante, M.; Riechmann, J.L.; Nomura, T.; et al. Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Jia, C.; Zhao, S.; Bao, T.; Zhao, P.; Peng, K.; Guo, Q.; Gao, X.; Qin, J. Tomato BZR/BES transcription factor SlBZR1 positively regulates BR signaling and salt stress tolerance in tomato and Arabidopsis. Plant Sci. 2021, 302, 110719. [Google Scholar] [CrossRef]
- Dong, N.Y.; Liu, D.; Zhang, X.; Yu, Z.; Huang, W.; Liu, J.; Yang, Y.; Meng, W.; Niu, M. Regulation of Brassinosteroid Signaling and Salt Resistance by SERK2 and Potential Utilization for Crop Improvement in Rice. Front. Plant Sci. 2020, 11, 621859. [Google Scholar] [CrossRef]
- Shang, Y.; Yang, D.; Ha, Y.; Shin, H.-Y.; Nam, K.H. RPK1 and BAK1 sequentially form complexes with OST1 to regulate ABA-induced stomatal closure. J. Exp. Bot. 2019, 71, 1491–1502. [Google Scholar] [CrossRef]
- Hu, Y.; Yu, D. BRASSINOSTEROID INSENSITIVE2 Interacts with ABSCISIC ACID INSENSITIVE5 to Mediate the Antagonism of Brassinosteroids to Abscisic Acid during Seed Germination in Arabidopsis. Plant Cell 2014, 26, 4394–4408. [Google Scholar] [CrossRef] [Green Version]
- Cai, Z.L.; Liu, J.; Wang, H.; Yang, C.; Chen, Y.; Li, Y.; Pan, S.; Dong, R.; Tang, G.; Barajas-Lopez, J.; et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 9651–9656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Bai, Y.; Shang, J.; Xin, R.; Tang, W. The antagonistic regulation of abscisic acid-inhibited root growth by brassinosteroids is partially mediated via direct suppression of ABSCISIC ACID INSENSITIVE 5 expression by BRASSINAZOLE RESISTANT 1. Plant Cell Environ. 2016, 39, 1994–2003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Guan, L.; Sun, Y.; Zhu, Y.; Liu, L.; Lu, R.; Jiang, M.; Tan, M.; Zhang, A. Calcium and ZmCCaMK are involved in brassinosteroid-induced antioxidant defense in maize leaves. Plant Cell Physiol. 2015, 56, 883–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Mao, T. Understanding the functions and mechanisms of plant cytoskeleton in response to environmental signals. Curr. Opin. Plant Biol. 2019, 52, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lin, F.; Mao, T.; Nie, J.; Yan, M.; Yuan, M.; Zhang, W. Phosphatidic Acid Regulates Microtubule Organization by Interacting with MAP65-1 in Response to Salt Stress in Arabidopsis. Plant Cell 2012, 24, 4555–4576. [Google Scholar] [CrossRef] [Green Version]
- Lian, N.; Wang, X.; Jing, Y.; Lin, J. Regulation of cytoskeleton-associated protein activities: Linking cellular signals to plant cytoskeletal function. J. Integr. Plant Biol. 2021, 63, 241–250. [Google Scholar] [CrossRef]
- Wang, C.Z.; Huang, R.D. Cytoskeleton and plant salt stress tolerance. Plant Signal Behav. 2014, 6, 29–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.; Han, R. Microtubule organization defects in Arabidopsis thaliana. Plant Biol. 2020, 22, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.G.; Zhu, D.Z.; Chen, G.H.; Gao, X.-Q.; Zhang, X.S. Disrupted actin dynamics trigger an increment in the reactive oxygen species levels in the Arabidopsis root under salt stress. Plant Cell Rep. 2012, 31, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, W.; Guo, Y. Arabidopsis SOS3 plays an important role in salt tolerance by mediating calcium-dependent microfilament reorganization. Plant Cell Rep. 2013, 32, 139–148. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, Z.; Guo, G.; Guo, Y. Microfilament Dynamics is Required for Root Growth under Alkaline Stress in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 952–958. [Google Scholar] [CrossRef]
- Wang, C.; Li, J.; Yuan, M. Salt Tolerance Requires Cortical Microtubule Reorganization in Arabidopsis. Plant Cell Physiol. 2007, 48, 1534–1547. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Zhang, L.; Yuan, M.; Ge, Y.; Liu, Y.; Fan, J.; Ruan, Y.; Cui, Z.; Tong, S.; Zhang, S. The microfilament cytoskeleton plays a vital role in salt and osmotic stress tolerance in Arabidopsis. Plant Biol. 2009, 12, 70–78. [Google Scholar] [CrossRef]
- Qian, D.; Xiang, Y. Actin Cytoskeleton as Actor in Upstream and Downstream of Calcium Signaling in Plant Cells. Int. J. Mol. Sci. 2019, 20, 1403. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Pan, Z.; Zhang, Y.; Qu, X.; Yang, Y.; Jiang, X.; Huang, S.; Yuan, M.; Schumaker, K.S.; Guo, Y. The Actin-Related Protein2/3 Complex Regulates Mitochondrial-Associated Calcium Signaling during Salt Stress in Arabidopsis. Plant Cell 2013, 25, 4544–4559. [Google Scholar] [CrossRef] [Green Version]
- Endler, A.; Kesten, C.; Schneider, R.; Zhang, Y.; Ivakov, A.; Froehlich, A.; Funke, N.; Persson, S. A Mechanism for Sustained Cellulose Synthesis during Salt Stress. Cell 2015, 162, 1353–1364. [Google Scholar] [CrossRef] [Green Version]
- Monniaux, M.; Hay, A. Cells, walls, and endless forms. Curr. Opin. Plant Biol. 2016, 34, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Tang, R.; Zhang, X.; Luan, S.; Yu, F. FERONIA Receptor Kinase at the Crossroads of Hormone Signaling and Stress Responses. Plant Cell Physiol. 2017, 58, 1143–1150. [Google Scholar] [CrossRef]
- Zhao, C.; Zayed, O.; Yu, Z.; Jiang, W.; Zhu, P.; Hsu, C.-C.; Zhang, L.; Tao, W.A.; Lozano-Durán, R.; Zhu, J.-K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Lee, C.; Ye, Z.-H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010, 15, 625–632. [Google Scholar] [CrossRef]
- Du, Q.; Wang, H. The role of HD-ZIP III transcription factors and miR165/166 in vascular development and secondary cell wall formation. Plant Signal. Behav. 2015, 10, e1078955. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Ye, Z.-H. MYB46 and MYB83 Bind to the SMRE Sites and Directly Activate a Suite of Transcription Factors and Secondary Wall Biosynthetic Genes. Plant Cell Physiol. 2012, 53, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Endler, A.; Persson, S. Cellulose Synthases and Synthesis in Arabidopsis. Mol. Plant 2011, 4, 199–211. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Hart, B.E.; Khan, G.A.; Cruz, E.R.; Persson, S.; Wallace, I.S. Associations between phytohormones and cellulose biosynthesis in land plants. Ann. Bot. 2020, 126, 807–824. [Google Scholar] [CrossRef]
- Liu, C.Y.; Rao, X.L.; Li, L.G.; Dixon, R.A. Abscisic acid regulates secondary cell-wall formation and lignin deposition in Arabidopsis thaliana through phosphorylation of NST1. Proc. Natl. Acad. Sci. USA 2021, 118, e2010911118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endler, A.; Schneider, R.; Kesten, C.; Lampugnani, E.R.; Persson, S. The cellulose synthase companion proteins act non-redundantly with CELLULOSE SYNTHASE INTERACTING1/POM2 and CELLULOSE SYNTHASE 6. Plant Signal. Behav. 2016, 11, e1135281. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Schneider, R.; Kesten, C.; Zhang, Y.; Somssich, M.; Zhang, Y.; Fernie, A.R.; Persson, S. Cellulose-Microtubule Uncoupling Proteins Prevent Lateral Displacement of Microtubules during Cellulose Synthesis in Arabidopsis. Dev. Cell 2016, 38, 305–315. [Google Scholar] [CrossRef] [Green Version]
- Kesten, C.; Wallmann, A.; Schneider, R.; McFarlane, H.E.; Diehl, A.; Khan, G.A.; Van Rossum, B.-J.; Lampugnani, E.R.; Szymanski, W.G.; Cremer, N.; et al. The companion of cellulose synthase 1 confers salt tolerance through a Tau-like mechanism in plants. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Chun, H.J.; Baek, D.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Yun, D.-J.; Hong, Y.-S.; Kim, M.C. Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress. Plant Signal. Behav. 2019, 14, 1625697. [Google Scholar] [CrossRef]
- Yan, J.; Liu, Y.; Yang, L.; He, H.; Huang, Y.; Fang, L.; Scheller, H.V.; Jiang, M.; Zhang, A. Cell wall β-1,4-galactan regulated by the BPC1/BPC2-GALS1 module aggravates salt sensitivity in Arabidopsis thaliana. Mol. Plant 2021, 14, 411–425. [Google Scholar] [CrossRef]
- Geng, G.; Lv, C.; Stevanato, P.; Li, R.; Liu, H.; Yu, L.; Wang, Y. Transcriptome Analysis of Salt-Sensitive and Tolerant Genotypes Reveals Salt-Tolerance Metabolic Pathways in Sugar Beet. Int. J. Mol. Sci. 2019, 20, 5910. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.; Joshi, M.; Joshi, V. Transcriptomic Analysis of Short-Term Salt Stress Response in Watermelon Seedlings. Int. J. Mol. Sci. 2020, 21, 6036. [Google Scholar] [CrossRef]
- Xiong, Y.; Yan, H.; Liang, H.; Zhang, Y.; Guo, B.; Niu, M.; Jian, S.; Ren, H.; Zhang, X.; Li, Y.; et al. RNA-Seq analysis of Clerodendrum inerme (L.) roots in response to salt stress. BMC Genom. 2019, 20, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Wang, Z.; Zhang, Q.; Ci, H.; Wang, P.; Yu, L.; Jia, G. Genome-wide transcriptome analysis of the salt stress tolerance mechanism in Rosa chinensis. PLoS ONE 2018, 13, e0200938. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.-Q.; Gao, Q. Comparative physiological and biochemical mechanisms of salt tolerance in five contrasting highland quinoa cultivars. BMC Plant Biol. 2020, 20, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. https://doi.org/10.3390/ijms22094609
Zhao S, Zhang Q, Liu M, Zhou H, Ma C, Wang P. Regulation of Plant Responses to Salt Stress. International Journal of Molecular Sciences. 2021; 22(9):4609. https://doi.org/10.3390/ijms22094609
Chicago/Turabian StyleZhao, Shuangshuang, Qikun Zhang, Mingyue Liu, Huapeng Zhou, Changle Ma, and Pingping Wang. 2021. "Regulation of Plant Responses to Salt Stress" International Journal of Molecular Sciences 22, no. 9: 4609. https://doi.org/10.3390/ijms22094609