Plasmid Curing and Exchange Using a Novel Counter-Selectable Marker Based on Unnatural Amino Acid Incorporation at a Sense Codon
Abstract
:1. Introduction
2. Results and Discussion
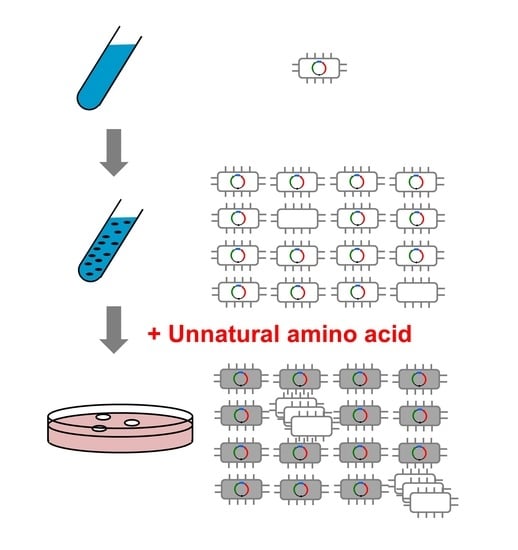
2.1. Plasmid Curing
2.2. Plasmid Exchange
3. Materials and Methods
3.1. Bacterial Strain, Culture, and Plasmid Transfection
3.2. Plasmid Curing
3.3. Plasmid Exchange
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reyrat, J.-M.; Pelicic, V.; Gicquel, B. Counterselectable markers: Untapped tools for bacterial genetics and pathogenesis. Infect. Immun. 1998, 66, 4011–4017. [Google Scholar] [CrossRef] [PubMed]
- Ried, J.L.; Collmer, A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in Gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 1987, 57, 239–246. [Google Scholar] [CrossRef]
- Blomfield, I.C.; Vaughn, V.; Rest, R.F.; Eisenstein, B.I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 1991, 5, 1447–1457. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Thomason, L.C.; Sawitzke, J.A.; Costantino, N.; Court, D.I. Positive and negative selection using the tetA-sacB cassette: Recombineering and P1 transduction in Escherichia coli. Nucl. Acids Res. 2013, 41, e204. [Google Scholar] [CrossRef] [PubMed]
- Ambrosis, N.; Fernández, J.; Sisti, F. Counter-selection method for markerless allelic exchange in Bordetella Bronchiseptica based on sacB gene from Bacillus subtilis. Curr. Protoc. Microbiol. 2020, 59, e125. [Google Scholar] [CrossRef]
- Pierce, J.C.; Sauer, B.; Sternberg, N. A positive selection vector for cloning high molecular weight DNA by the bacteriophage P1 system: Improved cloning efficacy. Proc. Natl. Acad. Sci. USA 1992, 89, 2056–2060. [Google Scholar] [CrossRef] [Green Version]
- Bernard, P.; Gabant, P.E.; Bahassi, M.; Couturier, M. Positive-selection vectors using the F plasmid ccdB killer gene. Gene 1994, 148, 71–74. [Google Scholar] [CrossRef]
- Hynes, M.F.; Quandt, J.; O’Connell, M.P.; Pühler, A. Direct selection for curing and deletion of Rhizobium plasmids using transposons carrying the Bacillus subtilis sacB gene. Gene 1989, 78, 111–120. [Google Scholar]
- Stojiljkovic, I.; Trgovcevic, Z.; Salaj-Smic, E. Tn5-rpsL: A new derivative of transposon Tn5 useful in plasmide curing. Gene 1991, 99, 101–104. [Google Scholar] [CrossRef]
- Lauritsen, I.; Kim, S.H.; Porse, A.; Nørholm, M.H.H. Standardized Cloning and Curing of Plasmids. Methods Mol. Biol. 2018, 1772, 469–476. [Google Scholar]
- Volke, D.C.; Friis, L.; Wirth, N.T.; Turlin, J.; Nikel, P.I. Synthetic control of plasmid replication enables target- and self-curing of vectors and expedites genome engineering of Pseudomonas putida. Metab. Eng. Commun. 2020, 10, e00126. [Google Scholar] [CrossRef]
- Raman, S.; Rogers, J.K.; Taylor, N.D.; Church, G.M. Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. USA 2014, 111, 17803–17808. [Google Scholar] [CrossRef] [Green Version]
- Schlegel, S.; Genevaux, P.; de Gier, J.W. De-convoluting the genetic adaptations of E. coli C41 (DE3) in real time reveals how alleviating protein production stress improves yields. Cell Rep. 2015, 10, 1758–1766. [Google Scholar] [PubMed] [Green Version]
- Dean, D. A plasmid cloning vector for the direct selection of strains carrying recombinant plasmids. Gene 1981, 15, 99–102. [Google Scholar] [CrossRef]
- Heermann, R.; Zeppenfeld, T.; Jung, K. Simple generation of site-directed point mutations in the Escherichia coli chromosome using Red®/ET® Recombination. Microb. Cell Fact. 2008, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; Wang, G.; Wang, Z.; Fu, J.; Chen, T.; Zhao, X. Establishment of a markerless mutation delivery system in Bacillus subtilis stimulated by a double-strand break in the chromosome. PLoS ONE 2013, 8, e81370. [Google Scholar] [CrossRef] [Green Version]
- Stringer, A.M.; Singh, N.; Yermakova, A.; Petrone, B.L.; Amarasinghe, J.J.; Reyes-Diaz, L.; Mantix, N.J.; Wade, J.T. FRUIT, a Scar-Free System for Targeted Chromosomal Mutagenesis, Epitope Tagging, and Promoter Replacement in Escherichia coli and Salmonella enterica. PLoS ONE 2012, 7, e44841. [Google Scholar] [CrossRef]
- Koropatkin, N.M.; Martens, E.C.; Gordon, J.I.; Smith, T.J. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 2008, 16, 1105–1115. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.Z. mazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis. Nucl. Acids Res. 2006, 34, e71. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yan, X.; Shen, W.; Shen, Y.; Zhang, J.; Li, S. Efficient and precise construction of markerless manipulations in the Bacillus subtilis genome. J. Microbiol. Biotechnol. 2010, 20, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinmetz, M.; Lecoq, D.; Djemia, H.B.; Gay, P. Analyse génétique de sacB, gène de structure d’une enzyme secrétée, la lévane-saccharase de Bacillus subtilis Marburg. Mol. Gen. Genet. 1983, 191, 138–144. [Google Scholar] [CrossRef]
- Jäger, W.; Schäfer, A.; Pühler, A.; Labes, G.; Wohlleben, W. Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J. Bacteriol. 1992, 174, 5462–5465. [Google Scholar] [CrossRef] [Green Version]
- Barrett, A.R.; Kang, Y.; Inamasu, K.S.; Son, M.S.; Vukovich, J.M.; Hoang, T.T. Genetic tools for allelic replacement in Burkholderia species. Appl. Environ. Microbiol. 2008, 74, 4498–4508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kast, P. PKSS—A second-generation general purpose cloning vector for efficient positive selection of recombinant clones. Gene 1994, 138, 109–114. [Google Scholar] [CrossRef]
- Miyazaki, K. Molecular engineering of a PheS counterselection marker for improved operating efficiency in Escherichia coli. Biotechniques 2015, 58, 86–88. [Google Scholar] [CrossRef]
- Xie, Z.; Okinaga, T.; Qi, F.; Zhang, Z.; Merritt, J. Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl. Environ. Microbiol. 2011, 77, 8025–8033. [Google Scholar] [CrossRef] [Green Version]
- Argov, T.; Rabinovich, L.; Sigal, N.; Herskovits, A.A. An effective counterselection system for Listeria monocytogenes and its use to characterize the monocin genomic region of strain 10403S. Appl. Environ. Microbiol. 2017, 83, e02927-16. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, M.; Yokoe, S.; Kato, S.; Hori, K. Efficient counterselection for Methylococcus capsulatus (Bath) by using amutated pheS gene. Appl. Environ. Microbiol. 2018, 84, e01875-1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, Y. An Unnatural Amino Acid-Regulated Growth Controller Based on Informational Disturbance. Life 2021, 11, 920. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, T.; Ishii, R.; Fukunaga, R.; Kobayashi, T.; Sakamoto, K.; Yokoyama, S. Multistep engineering of pyrrolysyl-tRNAsynthetase to genetically encode Nε-(o-Azidobenzyloxycarbonyl)lysine for site-specific protein modification. Chem. Biol. 2008, 15, 1187–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, W.; Tharp, J.M.; Liu, W.R. Pyrrolysyl-tRNA Synthetase: An ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta 2014, 1844, 1059–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambrogelly, A.; Gundllapalli, S.; Herring, S.; Polycarpo, C.; Frauer, C.; Söll, D. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl. Acad. Sci. USA 2007, 104, 3141–3146. [Google Scholar] [CrossRef] [Green Version]
- Chin, J.W. Expanding and reprogramming the genetic code. Nature 2017, 550, 53–60. [Google Scholar] [CrossRef]
- Shankar, R.; Selvamani, V.; Telaar, M.; Friehs, K.; Flaschel, E. Antibiotic-free segregational plasmid stabilization in Escherichia coli owing to the knockout of triosephosphate isomerase (tpiA). Microb. Cell Fact. 2014, 13, 58. [Google Scholar]
- Clark-Curtiss, J.E.; Curtiss III, R. Salmonella vaccines: Conduits for protective antigens. J. Immunol. 2018, 200, 39–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luke, J.; Carnes, A.E.; Hodgson, C.P.; Williams, J.A. Improved antibiotic-free DNA vaccine vectors utilizing a novel RNA basedplasmid selection system. Vaccine 2009, 27, 6454–6459. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, K.; Kelly, S.M.; Curtiss, R., III. Construction of an Asd+ expression-cloning vector: Stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat. Biotechnol. 1988, 6, 693–697. [Google Scholar] [CrossRef]
- Rose, R.E. The nucleotide sequence of pACYC184. Nucl. Acids Res. 1988, 16, 355. [Google Scholar] [CrossRef]
- Minaba, M.; Kato, Y. High-yield, zero-leakage expression system with a translational switch using site-specifc unnatural amino acid incorporation. Appl. Environ. Microbiol. 2014, 80, 1718–1725. [Google Scholar] [CrossRef]
- Saïda, F.; Uzan, M.; Odaert, B.; Bontems, F. Expression of highly toxicgenes in E. coli: Special strategies and genetic tools. Curr. Protein Pept. Sci. 2006, 7, 47–56. [Google Scholar] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kato, Y. Plasmid Curing and Exchange Using a Novel Counter-Selectable Marker Based on Unnatural Amino Acid Incorporation at a Sense Codon. Int. J. Mol. Sci. 2021, 22, 11482. https://doi.org/10.3390/ijms222111482
Kato Y. Plasmid Curing and Exchange Using a Novel Counter-Selectable Marker Based on Unnatural Amino Acid Incorporation at a Sense Codon. International Journal of Molecular Sciences. 2021; 22(21):11482. https://doi.org/10.3390/ijms222111482
Chicago/Turabian StyleKato, Yusuke. 2021. "Plasmid Curing and Exchange Using a Novel Counter-Selectable Marker Based on Unnatural Amino Acid Incorporation at a Sense Codon" International Journal of Molecular Sciences 22, no. 21: 11482. https://doi.org/10.3390/ijms222111482