Genome-Wide Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Petal Pigment Synthesis in Liriodendron
Abstract
:1. Introduction
2. Results
2.1. Identification of Genes and Prediction of Encoded Proteins of the R2R3-MYB Family
2.2. Phylogenetic Analysis of R2R3-MYB Family Genes in L. chinense
2.3. Gene Structure Analysis of R2R3-MYBs
2.4. Conserved Motifs within R2R3-MYB Proteins
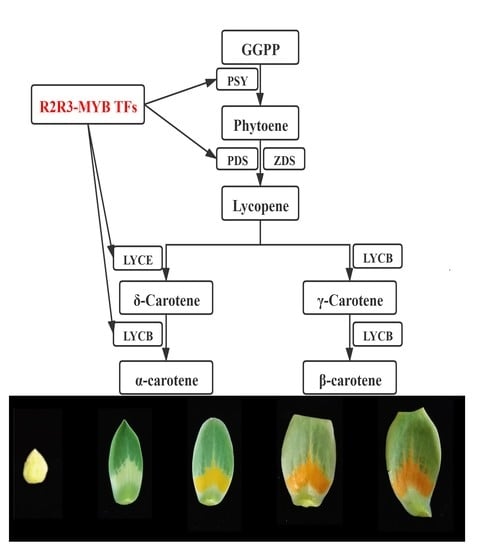
2.5. Dynamic Changes of Pigment during Petal Development
2.6. Prediction of Cis-Acting Elements in the Promoters of 16 R2R3-MYB Genes
2.7. RT-qPCR Assays of Twelve R2R3-MYB Genes across Five Stages of Petal Development in Liriodendron
3. Discussion
3.1. R2R3-MYB TFs Modulate Carotenoid Accumulation
3.2. Supplement of R2R3-MYB Members and Functional Modules between L. chinense and A. thaliana
3.3. ABA Regulates the Accumulation of Pigments
4. Materials and Methods
4.1. Plant Materials
4.2. Gene Identification and Coding Protein Prediction of L. chinense MYB Superfamily Members
4.3. Multiple Alignment and Phylogenetic Analysis
4.4. Gene Structure and Conserved Motif Analysis
4.5. Prediction of Cis-Acting Elements in the Promoters of Lcmybs
4.6. RNA Isolation and RT-qPCR Assays
4.7. Quantification of Photosynthetic Pigments and Carotenoids
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends. Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Qing, J.; Dawei, W.; Jun, Z.; Yulan, X.; Bingqi, S.; Fan, Z. Genome-wide characterization and expression analyses of the MYB superfamily genes during developmental stages in Chinese jujube. PeerJ 2019, 7, e6353. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.; Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q.; et al. The MYB Transcription Factor Superfamily of Arabidopsis: Expression Analysis and Phylogenetic Comparison with the Rice MYB Family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Yang, K.; Li, Y.; Wang, S.; Xu, X.; Sun, H.; Zhao, H.; Li, X.; Gao, Z. Genome-wide identification and expression analysis of the MYB transcription factor in moso bamboo (Phyllostachys edulis). PeerJ 2019, 6, e6242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Ma, R.; Xu, J.; Yan, J.; Guo, L.; Song, J.; Feng, R.; Yu, M. Genome-wide identification and classification of MYB superfamily genes in peach. PLoS ONE 2018, 13, e0199192. [Google Scholar] [CrossRef] [PubMed]
- Mmadi, M.A.; Dossa, K.; Wang, L.; Zhou, R.; Wang, Y.; Cisse, N.; Sy, M.O.; Zhang, X. Functional Characterization of the Versatile MYB Gene Family Uncovered Their Important Roles in Plant Development and Responses to Drought and Waterlogging in Sesame. Genes 2017, 8, 362. [Google Scholar] [CrossRef] [Green Version]
- Grotewold, E.; Drummond, B.J.; Bowen, B.; Peterson, T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 1994, 76, 543–553. [Google Scholar] [CrossRef]
- Oppenheimer, D.G.; Herman, P.L.; Sivakumaran, S.; Esch, J.; Marks, M.D. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 1991, 67, 483–493. [Google Scholar] [CrossRef]
- Liu, H.; Ma, J.; Li, H. Transcriptomic and microstructural analyses in Liriodendron tulipifera Linn. reveal candidate genes involved in nectary development and nectar secretion. BMC Plant Biol. 2019, 19, 531. [Google Scholar] [CrossRef] [Green Version]
- Baumann, K.; Perez-Rodriguez, M.; Bradley, D.; Venail, J.; Bailey, P.; Jin, H.; Koes, R.; Roberts, K.; Martin, C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 2007, 134, 1691–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urao, T.; Yamaguchi-Shinozaki, K.; Urao, S. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 1993, 5, 1529–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Klessig, D.F. Isolation and characterization of a tobacco mosaic virus-inducible myb oncogene homolog from tobacco. Proc. Natl. Acad. Sci. USA 1996, 93, 14972–14977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiou, C.Y.; Yeh, K.W. Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium Gower Ramsey. Plant Mol. Biol. 2008, 66, 379–388. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Wu, J.; Xue, Y.; Zhao, W.; Li, R.; Zhang, L. The novel gene BrMYB2, located on chromosome A07, with a short intron 1 controls the purple-head trait of Chinese cabbage (Brassica rapa L.). Hortic. Res. 2020, 7, 1–19. [Google Scholar] [CrossRef]
- Hao, Z.; Liu, S.; Hu, L.; Shi, J.; Chen, J. Transcriptome analysis and metabolic profiling reveal the key role of carotenoids in the petal coloration of Liriodendron tulipifera. Hortic. Res. 2020, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mol, J.; Grotewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 3, 212–217. [Google Scholar] [CrossRef]
- Bolger, M.E.; Weisshaar, B.; Scholz, U.; Stein, N.; Usadel, B.; Mayer, K.F. Plant genome sequencing—applications for crop improvement. Curr. Opin. Biotechnol. 2014, 26, 31–37. [Google Scholar] [CrossRef]
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y.; et al. Liriodendron genome sheds light on angiosperm phylogeny and species–pair differentiation. Nat. Plants 2019, 5, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Chhon, S.; Jeon, J.; Kim, J.; Park, S.U. Accumulation of Anthocyanins through Overexpression of AtPAP1 in Solanum nigrum Lin. (Black Nightshade). Biomolecules 2020, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Pandey, A.; Misra, P.; Trivedi, P.K. Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 2015, 34, 1515–1528. [Google Scholar] [CrossRef]
- Pandey, A.; Misra, P.J.; Bhambhani, S.; Bhatia, C.; Trivedi, P.K. Expression of Arabidopsis MYB transcription factor, AtMYB111, in tobacco requires light to modulate flavonol content. Sci. Rep. 2014, 4, 5018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.-L.; Tang, Y.; Zhang, K.-X.; Li, F.-L.; Yang, P.-Y.; Tang, Y.-X.; Wu, Y.-M.; Shao, J.-R. Identification of TT2 gene from floral transcriptome in Fagopyrum tataricum. Food Res. Int. 2013, 54, 1331–1333. [Google Scholar] [CrossRef]
- Hou, X.-J.; Li, S.-B.; Liu, S.-R.; Hu, C.-G.; Zhang, J.-Z. Genome-Wide Classification and Evolutionary and Expression Analyses of Citrus MYB Transcription Factor Families in Sweet Orange. PLoS ONE 2014, 9, e112375. [Google Scholar] [CrossRef]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and Diversification of the Populus R2R3-MYB Family of Transcription Factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Peng, R.; Tian, Y.; Han, H.; Xu, J.; Yao, Q. Genome-Wide Identification and Analysis of the MYB Transcription Factor Superfamily in Solanum lycopersicum. Plant Cell Physiol. 2016, 57, 1657–1677. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Xue, C.; Li, J.; Qiao, X.; Li, L.; Yu, L.; Huang, Y.; Wu, J. Genome-Wide Identification, Evolution and Functional Divergence of MYB Transcription Factors in Chinese White Pear (Pyrus bretschneideri). Plant Cell Physiol. 2016, 57, 824–847. [Google Scholar] [CrossRef] [Green Version]
- Nesi, N.; Jond, C.; Debeaujon, I.; Caboche, M.; Lepiniec, L. The Arabidopsis TT2 Gene Encodes an R2R3 MYB Domain Protein That Acts as a Key Determinant for Proanthocyanidin Accumulation in Developing Seed. Plant Cell 2001, 13, 2099–2114. [Google Scholar] [CrossRef] [Green Version]
- Zhai, R.; Wang, Z.; Zhang, S.; Meng, G.; Song, L.; Wang, Z.; Li, P.; Ma, F.; Xu, L. Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.). J. Exp. Bot. 2016, 67, 1275–1284. [Google Scholar] [CrossRef] [Green Version]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.H.; Espley, R.; Allan, A.C. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef] [Green Version]
- Zhong, C.; Tang, Y.; Pang, B.; Li, X.; Yang, Y.; Deng, J.; Feng, C.; Li, L.; Ren, G.; Wang, Y.; et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 2020, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Luo, T.; Liu, C.; Wang, Y.; Yang, H.; Yang, W.; Zheng, L.; Xiao, X.; Zhang, M.; Xu, R.; et al. An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 2017, 216, 178–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apel, W.; Bock, R. Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. Plant Physiol. 2009, 151, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Xu, J.; Liu, Y.; Zhao, X.; Deng, X.; Guo, L.; Gu, J. A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J. Exp. Bot. 2007, 58, 4161–4171. [Google Scholar] [CrossRef] [PubMed]
- Castillejo, C.; Waurich, V.; Wagner, H.; Ramos, R.; Oiza, N.; Muñoz, P.; Triviño, J.C.; Caruana, J.; Liu, Z.; Cobo, N.; et al. Allelic Variation of MYB10 Is the Major Force Controlling Natural Variation in Skin and Flesh Color in Strawberry (Fragaria spp.). Fruit. Plant Cell 2020, 32, 3723–3749. [Google Scholar] [CrossRef]
- Li, B.-J.; Zheng, B.-Q.; Wang, J.-Y.; Tsai, W.-C.; Lu, H.-C.; Zou, L.-H.; Wan, X.; Zhang, D.-Y.; Qiao, H.-J.; Liu, Z.-J.; et al. New insight into the molecular mechanism of colour differentiation among floral segments in orchids. Commun. Biol. 2020, 3, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kay, Q.O.N.; Daoud, H.S.; Stirton, C.H. Pigment distribution, light reflection and cell structure in petals. Bot. J. Linn. Soc. 1981, 83, 57–83. [Google Scholar] [CrossRef]
- Jing, Y. Study on genetic variation of perianth pigment and quantitative characters of flower and fruit in Liriodendron. Master’s Thesis, Nanjing Forestry University, Nanjing, China, 2005. [Google Scholar]
- Luan, Y.; Fu, X.; Lu, P.; Grierson, D.; Xu, C. Molecular Mechanisms Determining the Differential Accumulation of Carotenoids in Plant Species and Varieties. Crit. Rev. Plant Sci. 2020, 39, 125–139. [Google Scholar] [CrossRef]
- Meng, Y.; Wang, Z.; Wang, Y.; Wang, C.; Zhu, B.; Liu, H.; Ji, W.; Wen, J.; Chu, C.; Tadege, M.; et al. The MYB Activator WHITE PETAL1 Associates with MtTT8 and MtWD40-1 to Regulate Carotenoid-Derived Flower Pigmentation in Medicago truncatula. Plant Cell 2019, 31, 2751–2767. [Google Scholar] [CrossRef] [Green Version]
- Sagawa, J.M.; Stanley, L.E.; LaFountain, A.M.; Frank, H.A.; Liu, C.; Yuan, Y. An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 2016, 209, 1049–1057. [Google Scholar] [CrossRef]
- Wu, M.; Xu, X.; Hu, X.; Liu, Y.; Cao, H.; Chan, H.; Gong, Z.; Yuan, Y.; Luo, Y.; Feng, B.; et al. SlMYB72 Regulates the Metabolism of Chlorophylls, Carotenoids, and Flavonoids in Tomato Fruit. Plant Physiol. 2020, 183, 854–868. [Google Scholar] [CrossRef]
- Hao, Z.; Zong, Y.; Liu, H.; Tu, Z.; Li, H. Cloning, Characterization and Functional Analysis of the LtuPTOX Gene, a Homologue of Arabidopsis thaliana IMMUTANS Derived from Liriodendron tulipifera. Genes 2019, 10, 878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Zhu, S.; Zhu, L.; Wang, D.; Liu, Y.; Liu, S.; Zhang, J.; Hao, Z.; Lu, Y.; Cheng, T.; et al. Characterization of the Liriodendron Chinense MYB Gene Family and Its Role in Abiotic Stress Response. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Browse, M.J. MYB108 Acts Together with MYB24 to Regulate Jasmonate-Mediated Stamen Maturation in Arabidopsis. Plant Physiol. 2009, 149, 851–862. [Google Scholar] [CrossRef] [Green Version]
- Gigolashvili, T.; Berger, B.; Mock, H.-P.; Müller, C.; Weisshaar, B.; Flügge, U.-I. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007, 50, 886–901. [Google Scholar] [CrossRef] [Green Version]
- Stracke, R.; Holtgräwe, D.; Schneider, J.; Pucker, B.; Sörensen, T.R.; Weisshaar, B. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol. 2014, 14, 249. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Yang, S.-S.; Liang, Z.; Feng, B.-R.; Liu, L.; Huang, Y.-B.; Tang, Y.-X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Chen, J.; Gu, H.; Cheng, D.; Zhang, W.; Zhang, Y. Influencing factors and improvement measures of grape fruit coloring. Sino-Overseas Grapevine Wine 2016, 5, 124–127. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [Green Version]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Yu, M.; Liu, H.; Wang, J. Effects of ABA on expression of genes related to anthocyanin biosynthesis in grapevine. J. Fruit Sci. 2012, 29, 29–35, CNKI:SUN:GSKK.0.2012-01-007. [Google Scholar]
- Jeong, S.; Goto-Yamamoto, N.; Kobayashi, S.; Esaka, M. Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci. 2004, 167, 247–252. [Google Scholar] [CrossRef]
- Katayama-Ikegami, A.; Sakamoto, T.; Shibuya, K.; Katayama, T.; Gao-Takai, M. Effects of Abscisic Acid Treatment on Berry Coloration and Expression of Flavonoid Biosynthesis Genes in Grape. Am. J. Plant Sci. 2016, 07, 1325–1336. [Google Scholar] [CrossRef] [Green Version]
- Medina-Puche, L.; Cumplido-Laso, G.; Amil-Ruiz, F.; Hoffmann, T.; Ring, L.; Franco, A.R.; Caballero, J.L.; Schwab, W.; Blanco, J.M.; Portales, R.B. MYB10plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria×ananassa fruits. J. Exp. Bot. 2014, 65, 401–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Zhao, K.; Liu, L.; Zhang, K.; Yuan, H.; Liao, X.; Wang, Q.; Guo, X.; Li, F.; Li, T. A Role for PacMYBA in ABA-Regulated Anthocyanin Biosynthesis in Red-Colored Sweet Cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol. 2014, 55, 862–880. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-L.; Wang, Y.-X.; Li, H.; Liu, Z.-W.; Cui, X.; Zhuang, J. Two MYB transcription factors (CsMYB2 and CsMYB26) are involved in flavonoid biosynthesis in tea plant [Camellia sinensis (L.) O. Kuntze]. BMC Plant Biol. 2018, 18, 1–15. [Google Scholar] [CrossRef]
- Zhang, M.; Yuan, B.; Leng, P. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J. Exp. Bot. 2009, 60, 1579–1588. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Guo, S.; Ren, Y.; Zhang, H.; Gong, G.; Zhou, M.; Wang, G.; Zong, M.; He, H.; Liu, F.; et al. High-level expression of a novel chromoplast phosphate transporter ClPHT4;2 is required for flesh color development in watermelon. New Phytol. 2017, 213, 1208–1221. [Google Scholar] [CrossRef]
- Zaharah, S.S.; Singh, Z.; Symons, G.M.; Reid, J.B. Role of Brassinosteroids, Ethylene, Abscisic Acid, and Indole-3-Acetic Acid in Mango Fruit Ripening. J. Plant Growth Regul. 2012, 31, 363–372. [Google Scholar] [CrossRef]
- Barickman, T.C.; Kopsell, D.; Sams, C.E. Abscisic Acid Increases Carotenoid and Chlorophyll Concentrations in Leaves and Fruit of Two Tomato Genotypes. J. Am. Soc. Hortic. Sci. 2014, 139, 261–266. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sun, L.; Xie, L.; He, Y.; Luo, T.; Sheng, L.; Luo, Y.; Zeng, Y.; Xu, J.; Deng, X.; et al. Regulation of cuticle formation during fruit development and ripening in ‘Newhall’ navel orange (Citrus sinensis Osbeck) revealed by transcriptomic and metabolomic profiling. Plant Sci. 2016, 243, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vallabhaneni, R.; Wurtzel, E.T. PSY3, a New Member of the Phytoene Synthase Gene Family Conserved in the Poaceae and Regulator of Abiotic Stress-Induced Root Carotenogenesis. Plant Physiol. 2008, 146, 1333–1345. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Wang, N.; Cui, F.; Li, X.; Xiao, J.; Xiong, L. Characterization of the β-Carotene Hydroxylase Gene DSM2 Conferring Drought and Oxidative Stress Resistance by Increasing Xanthophylls and Abscisic Acid Synthesis in Rice. Plant Physiol. 2010, 154, 1304–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welsch, R.; Wust, F.; Bar, C.; Al-Babili, S.; Beyer, P. A Third Phytoene Synthase Is Devoted to Abiotic Stress-Induced Abscisic Acid Formation in Rice and Defines Functional Diversification of Phytoene Synthase Genes. Plant Physiol. 2008, 147, 367–380. [Google Scholar] [CrossRef] [Green Version]
- Kirik, V.; Kölle, K.; Miséra, S.; Bäumlein, H. Two novel MYB homologues with changed expression in late embryogenesis-defective Arabidopsis mutants. Plant Mol. Biol. 1998, 37, 819–827. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Chang, H.-S.; Gupta, R.; Wang, X.; Zhu, T.; Luan, S. Transcriptional Profiling Reveals Novel Interactions between Wounding, Pathogen, Abiotic Stress, and Hormonal Responses in Arabidopsis. Plant Physiol. 2002, 129, 661–677. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.; Nothnagel, E.A. Binding of Arabinogalactan Proteins by Yariv Phenylglycoside Triggers Wound-Like Responses in Arabidopsis Cell Cultures. Plant Physiol. 2004, 135, 1346–1366. [Google Scholar] [CrossRef] [Green Version]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.-J. Overexpression of AtMYB44 Enhances Stomatal Closure to Confer Abiotic Stress Tolerance in Transgenic Arabidopsis. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Yang, L.; Chen, X.; Li, L.; Guo, D.; Li, H.-H.; Zhang, B. Cloning and Characterization of the Promoter of the 9-cis-Epoxycarotenoid Dioxygenase Gene in Arachis hypogaea L. Biosci. Biotechnol. Biochem. 2009, 73, 2103–2106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, M.K.; Lindemose, S.; De Masi, F.; Reimer, J.J.; Nielsen, M.; Perera, V.; Workman, C.; Turck, F.; Grant, M.R.; Mundy, J.; et al. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 2013, 3, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Liang, G.; Yu, D. Activated Expression of WRKY57 Confers Drought Tolerance in Arabidopsis. Mol. Plant 2012, 5, 1375–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, Z.; Hao, Z.; Zhong, W.; Li, H. Identification of Suitable Reference Genes for RT-qPCR Assays in Liriodendron chinense (Hemsl.). Sarg. For. 2019, 10, 441. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwak, M.J.; Je, S.M.; Cheng, H.C.; Seo, S.M.; Park, J.H.; Baek, S.G.; Khaine, I.; Lee, T.; Jang, J.; Li, Y.; et al. Night Light-Adaptation Strategies for Photosynthetic Apparatus in Yellow-Poplar (Liriodendron tulipifera L.) Exposed to Artificial Night Lighting. Forests 2018, 9, 74. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Li, J. Study on the Changes of Carotenoid Compositions in Pepper (Capsicum annuum L.) Leaves under Low Temperature and Light and Its Relationship with the Tolerance of Varieties. Ph.D. Thesis, Gansu Agricultural University, Lanzhou, China, 2018. [Google Scholar]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Liu, H.; Hao, Z.; Zong, Y.; Xia, H.; Shen, Y.; Li, H. Genome-Wide Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Petal Pigment Synthesis in Liriodendron. Int. J. Mol. Sci. 2021, 22, 11291. https://doi.org/10.3390/ijms222011291
Yang L, Liu H, Hao Z, Zong Y, Xia H, Shen Y, Li H. Genome-Wide Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Petal Pigment Synthesis in Liriodendron. International Journal of Molecular Sciences. 2021; 22(20):11291. https://doi.org/10.3390/ijms222011291
Chicago/Turabian StyleYang, Lichun, Huanhuan Liu, Ziyuan Hao, Yaxian Zong, Hui Xia, Yufang Shen, and Huogen Li. 2021. "Genome-Wide Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Petal Pigment Synthesis in Liriodendron" International Journal of Molecular Sciences 22, no. 20: 11291. https://doi.org/10.3390/ijms222011291