Aurora B Tension Sensing Mechanisms in the Kinetochore Ensure Accurate Chromosome Segregation
Abstract
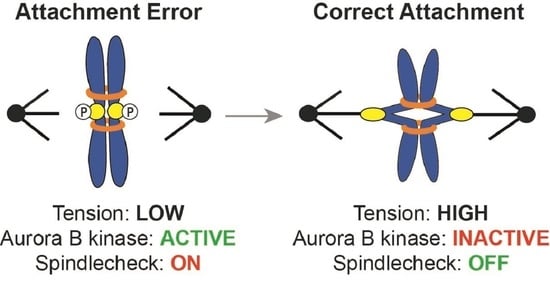
:1. Introduction
2. Tension Generation on Correct Attachments
3. Tension vs. Attachment: Activation of the Spindle Assembly Checkpoint
4. Where Is Tension Sensed? Inter-Kinetochore vs. Intra-Kinetochore Stretch
5. Responding to Tension: Aurora B Kinase
5.1. Aurora B and the Chromosome Passenger Complex
5.2. Targets and Consequences of Aurora B Phosphorylation
6. Aurora B Tension-Sensing Models
6.1. Spatial Separation Models
6.1.1. Spatial Separation: Dog Leash Model
6.1.2. Spatial Separation: Centromere Gradient Model
6.2. Tension-Sensitive Activation Models
6.3. Kinetochore Localization Model
6.4. Downstream Response Model
7. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hassold, T.; Hall, H.; Hunt, P. The origin of human aneuploidy: Where we have been, where we are going. Hum. Mol. Genet. 2007, 16, R203–R208. [Google Scholar] [CrossRef]
- Ben-David, U.; Amon, A. Context is everything: Aneuploidy in cancer. Nat. Rev. Genet. 2020, 21, 44–62. [Google Scholar] [CrossRef]
- Musacchio, A.; Desai, A. A Molecular view of kinetochore assembly and function. Biology 2017, 6, 5. [Google Scholar] [CrossRef] [Green Version]
- Winey, M.; Mamay, C.L.; O’Toole, E.T.; Mastronarde, D.N.; Giddings, T.H., Jr.; McDonald, K.L.; McIntosh, J.R. Three-dimensional ultrastructural analysis of the saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995, 129, 1601–1615. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, E.T.; Winey, M.; McIntosh, J.R. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast saccharomyces cerevisiae. Mol. Biol. Cell 1999, 10, 2017–2031. [Google Scholar] [CrossRef]
- McEwen, B.F.; Dong, Y. Contrasting models for kinetochore microtubule attachment in mammalian cells. Cell. Mol. Life Sci. 2010, 67, 2163–2172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassold, T.; Hunt, P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001, 2, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 2018, 33, 676–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.; Taunk, N.K. Chromosomal instability drives metastasis through a cytosolic dna response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakhoum, S.F.; Cantley, L.C. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 2018, 174, 1347–1360. [Google Scholar] [CrossRef] [Green Version]
- Etemad, B.; Kops, G.J. Attachment issues: Kinetochore transformations and spindle checkpoint silencing. Curr. Opin. Cell Biol. 2016, 39, 101–108. [Google Scholar] [CrossRef]
- Li, R.; Murray, A.W. Feedback Control of Mitosis in Budding Yeast. Cell 1991, 66, 519–531. [Google Scholar] [CrossRef]
- Hoyt, M.A.; Totis, L.; Roberts, B.T.S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 1991, 66, 507–517. [Google Scholar] [CrossRef]
- Hardwick, K.G.; Weiss, E.; Luca, F.C.; Winey, M.; Murray, A.W. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science 1996, 273, 953–956. [Google Scholar] [CrossRef] [Green Version]
- Hwang, L.H.; Lau, L.F.; Smith, D.L.; Mistrot, C.A.; Hardwick, K.G.; Hwang, E.S.; Amon, A.; Murray, A.W. Budding yeast Cdc20: A target of the spindle checkpoint. Science 1998, 279, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Lin, D.P.; Matsumoto, S.; Kitazono, A.; Matsumoto, T. Fission yeast Slp1: An effector of the Mad2-dependent spindle checkpoint. Science 1998, 279, 1045–1047. [Google Scholar] [CrossRef]
- Alfieri, C.; Chang, L.; Zhang, Z.; Yang, J.; Maslen, S.; Skehel, M.; Barford, D. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature 2016, 536, 431–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, M.; VanderLinden, R.; Weissmann, F.; Qiao, R.; Dube, P.; Brown, N.; Haselbach, D.; Zhang, W.; Sidhu, S.; Peters, J.; et al. Cryo-EM of mitotic checkpoint complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Mol. Cell 2016, 63, 593–607. [Google Scholar] [CrossRef] [Green Version]
- Lara-Gonzalez, P.; Pines, J.; Desai, A. Spindle assembly checkpoint activation and silencing at kinetochores. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Jin, F.; Wang, Y. The Signaling Network that Silences the Spindle Assembly Checkpoint upon the Establishment of Chromosome Bipolar Attachment. Proc. Natl. Acad. Sci. USA 2013, 110, 21036–21041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foley, E.A.; Kapoor, T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013, 14, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Nasmyth, K. How do so few control so many? Cell 2005, 120, 739–746. [Google Scholar] [CrossRef] [Green Version]
- Guacci, V.; Koshland, D.; Strunnikov, A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 1997, 91, 47–57. [Google Scholar] [CrossRef] [Green Version]
- Haering, C.H.; Farcas, A.; Arumugam, P.; Metson, J.; Nasmyth, K. The cohesin ring concatenates sister DNA molecules. Nature 2008, 454, 297–301. [Google Scholar] [CrossRef] [Green Version]
- Lengronne, A.; Katou, Y.; Mori, S.; Yokobayashi, S.; Kelly, G.P.; Itoh, T.; Watanabe, Y.; Shirahige, K.; Uhlmann, F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 2004, 430, 573–578. [Google Scholar] [CrossRef]
- Uhlmann, F.; Wernic, D.; Poupart, M.; Koonin, E.V.; Nasmyth, K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 2000, 103, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Nicklas, R.B. A quantitative study of chromosomal elasticity and its influence on chromosome movement. Chromosoma 1963, 14, 276–295. [Google Scholar] [CrossRef]
- Salmon, E.D.; Bloom, K. Tension sensors reveal how the kinetochore shares its load. Bioessays 2017, 39, 1600216. [Google Scholar] [CrossRef] [PubMed]
- Anna, A.Y.; Cane, S.; Maresca, T.J. Chromosome biorientation produces hundreds of piconewtons at a metazoan kinetochore. Nat. Commun. 2016, 7, 1–9. [Google Scholar]
- Suzuki, A.; Badger, B.L.; Haase, J.; Ohashi, T.; Erickson, H.P.; Salmon, E.D.; Bloom, K. How the kinetochore couples microtubule force and centromere stretch to move chromosomes. Nat. Cell Biol. 2016, 18, 382–392. [Google Scholar] [CrossRef] [Green Version]
- King, J.M.; Nicklas, R.B. Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J. Cell Sci. 2000, 113, 3815–3823. [Google Scholar] [CrossRef] [PubMed]
- Akiyoshi, B.; Sarangapani, K.K.; Powers, A.F.; Nelson, C.R.; Reichow, S.L.; Arellano-Santoyo, H.; Gonen, T.; Ranish, J.A.; Asbury, C.L.; Biggins, S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 2010, 468, 576–579. [Google Scholar] [CrossRef] [Green Version]
- Umbreit, N.T.; Gestaut, D.R.; Tien, J.F.; Vollmar, B.S.; Gonen, T.; Asbury, C.L.; Davis, T.N. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc. Natl. Acad. Sci. USA 2012, 109, 16113–16118. [Google Scholar] [CrossRef] [Green Version]
- Straight, A.F.; Marshall, W.F.; Sedat, J.W.; Murray, A.W. Mitosis in living budding yeast: Anaphase a but no metaphase plate. Science 1997, 277, 574–578. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Renda, F.; Zhang, H.; Gokden, A.; Wu, D.Z.; Chenoweth, D.M.; Khodjakov, A.; Lampson, M.A. Tension promotes kinetochore–microtubule release by aurora B kinase. J. Cell Biol. 2021, 220, e202007030. [Google Scholar] [CrossRef]
- Thomas, W.E.; Vogel, V.; Sokurenko, E. Biophysics of catch bonds. Annu. Rev. Biophys. 2008, 37, 399–416. [Google Scholar] [CrossRef]
- Willems, E.; Dedobbeleer, M.; Digregorio, M.; Lombard, A.; Lumapat, P.N.; Rogister, B. The functional diversity of aurora kinases: A comprehensive review. Cell Div. 2018, 13, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broad, A.J.; DeLuca, J.G. The right place at the right time: Aurora B kinase localization to centromeres and kinetochores. Essays Biochem. 2020, 64, 299–311. [Google Scholar]
- Nicklas, R.B.; Koch, C.A. CHROMOSOME MICROMANIPULATION: III. Spindle Fiber Tension and the Reorientation of Mal-Oriented Chromosomes. J. Cell Biol. 1969, 43, 40–50. [Google Scholar] [CrossRef] [Green Version]
- Rieder, C.L.; Cole, R.W.; Khodjakov, A.; Sluder, G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 1995, 130, 941–948. [Google Scholar] [CrossRef] [PubMed]
- DeLuca, K.F.; Lens, S.M.; DeLuca, J.G. Temporal changes in Hec1 phosphorylation control kinetochore–microtubule attachment stability during mitosis. J. Cell Sci. 2011, 124, 622–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaytsev, A.V.; Sundin, L.J.R.; DeLuca, K.F.; Grishchuk, E.L.; DeLuca, J.G. Accurate phosphoregulation of kinetochore–microtubule affinity requires unconstrained molecular interactions. J. Cell Biol. 2014, 206, 45–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biggins, S.; Murray, A.W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001, 15, 3118–3129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welburn, J.P.; Vleugel, M.; Liu, D.; Yates, J.R., III; Lampson, M.A.; Fukagawa, T.; Cheeseman, I.M. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 2010, 38, 383–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Sandri, B.J.; Tank, D.; McClellan, M.; Harasymiw, L.A.; Yang, Q.; Parker, L.L.; Gardner, M.K. A gradient in metaphase tension leads to a scaled cellular response in mitosis. Dev. Cell 2019, 49, 63–76. [Google Scholar] [CrossRef] [Green Version]
- Pinsky, B.A.; Biggins, S. The spindle checkpoint: Tension versus attachment. Trends Cell Biol. 2005, 15, 486–493. [Google Scholar] [CrossRef]
- Stern, B.M.; Murray, A.W. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 2001, 11, 1462–1467. [Google Scholar] [CrossRef] [Green Version]
- Henriques, A.C.; Silva, P.M.A.; Sarmento, B.; Bousbaa, H. Antagonizing the spindle assembly checkpoint silencing enhances paclitaxel and navitoclax-mediated apoptosis with distinct mechanistic. Sci. Rep. 2021, 11, 4139. [Google Scholar] [CrossRef]
- Taveras, C.; Liu, C.; Mao, Y. A tension-independent mechanism reduces aurora B-mediated phosphorylation upon microtubule capture by CENP-E at the kinetochore. Cell Cycle 2019, 18, 1349–1363. [Google Scholar] [CrossRef]
- Vallot, A.; Leontiou, I.; Cladière, D.; El Yakoubi, W.; Bolte, S.; Buffin, E.; Wassmann, K. Tension-induced error correction and not kinetochore attachment status activates the SAC in an Aurora-B/C-dependent manner in oocytes. Curr. Biol. 2018, 28, 130–139. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, P.; Zaytsev, A.V.; Godzi, M.; Ataullakhanov, F.I.; Grishchuk, E.L.; Stukenberg, P.T. The binding of borealin to microtubules underlies a tension independent kinetochore-microtubule error correction pathway. Nat. Commun. 2019, 10, 1–19. [Google Scholar] [CrossRef]
- Etemad, B.; Kuijt, T.E.; Kops, G.J. Kinetochore–microtubule attachment is sufficient to satisfy the human spindle assembly checkpoint. Nat. Commun. 2015, 6, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tauchman, E.C.; Boehm, F.J.; DeLuca, J.G. Stable kinetochore–microtubule attachment is sufficient to silence the spindle assembly checkpoint in human cells. Nat. Commun. 2015, 6, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, J.; Dumont, S. Mammalian kinetochores count attached microtubules in a sensitive and switch-like manner. J. Cell Biol. 2019, 218, 3583–3596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santaguida, S.; Vernieri, C.; Villa, F.; Ciliberto, A.; Musacchio, A. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 2011, 30, 1508–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, L.B.; Opoku, K.N.; Deng, Y.; Barber, A.; Littleton, A.J.; London, N.; Biggins, S.; Asbury, C.L. Autophosphorylation is sufficient to release Mps1 kinase from native kinetochores. Proc. Natl. Acad. Sci. USA 2019, 116, 17355–17360. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Vader, G.; Vromans, M.J.; Lampson, M.A.; Lens, S.M. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 2009, 323, 1350–1353. [Google Scholar] [CrossRef] [Green Version]
- Uchida, K.S.; Takagaki, K.; Kumada, K.; Hirayama, Y.; Noda, T.; Hirota, T. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 2009, 184, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Maresca, T.J.; Salmon, E.D. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 2009, 184, 373–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumont, S.; Salmon, E.D.; Mitchison, T.J. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science 2012, 337, 355–358. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Asthana, S.; Sorger, P.K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 2000, 101, 763–775. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Fuchs, J.; Loidl, J.; Nasmyth, K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000, 2, 492–499. [Google Scholar] [CrossRef]
- Goshima, G.; Yanagida, M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 2000, 100, 619–633. [Google Scholar] [CrossRef] [Green Version]
- Indjeian, V.B.; Murray, A.W. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr. Biol. 2007, 17, 1837–1846. [Google Scholar] [CrossRef] [Green Version]
- Bouck, D.C.; Bloom, K. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 2007, 17, 741–748. [Google Scholar] [CrossRef] [Green Version]
- Gay, G.; Courtheoux, T.; Reyes, C.; Tournier, S.; Gachet, Y. A stochastic model of kinetochore-microtubule attachment accurately describes fission yeast chromosome segregation. J. Cell Biol. 2012, 196, 757–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephens, A.D.; Haase, J.; Vicci, L.; Taylor, R.M.; Bloom, K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J. Cell Biol. 2011, 193, 1167–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephens, A.D.; Snider, C.E.; Haase, J.; Haggerty, R.A.; Vasquez, P.A.; Forest, M.G.; Bloom, K. Individual pericentromeres display coordinated motion and stretching in the yeast spindle. J. Cell Biol. 2013, 203, 407–416. [Google Scholar] [CrossRef]
- Lawrimore, J.; Bloom, K. The regulation of chromosome segregation via centromere loops. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 352–370. [Google Scholar] [CrossRef] [PubMed]
- Prota, A.E.; Danel, F.; Bachmann, F.; Bargsten, K.; Buey, R.M.; Pohlmann, J.; Reinelt, S.; Lane, H.; Steinmetz, M.O. The novel microtubule-destabilizing drug BAL27862 binds to the colchicine site of tubulin with distinct effects on microtubule organization. J. Mol. Biol. 2014, 426, 1848–1860. [Google Scholar] [CrossRef]
- Dudka, D.; Noatynska, A.; Smith, C.A.; Liaudet, N.; McAinsh, A.D.; Meraldi, P. Complete microtubule–kinetochore occupancy favours the segregation of merotelic attachments. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Nannas, N.J.; O’Toole, E.T.; Winey, M.; Murray, A.W. Chromosomal attachments set length and microtubule number in the saccharomyces cerevisiae mitotic spindle. Mol. Biol. Cell 2014, 25, 4034–4048. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; O’Quinn, R.P.; Pierce, H.L.; Joglekar, A.P.; Gall, W.E.; DeLuca, J.G.; Carroll, C.W.; Liu, S.; Yen, T.J.; McEwen, B.F. Protein architecture of the human kinetochore microtubule attachment site. Cell 2009, 137, 672–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, K.S.; Jo, M.; Nagasaka, K.; Takahashi, M.; Shindo, N.; Shibata, K.; Tanaka, K.; Masumoto, H.; Fukagawa, T.; Hirota, T. Kinetochore stretching-mediated rapid silencing of the spindle-assembly checkpoint required for failsafe chromosome segregation. Curr. Biol. 2021, 31, 1581–1591. [Google Scholar] [CrossRef]
- Roscioli, E.; Germanova, T.E.; Smith, C.A.; Embacher, P.A.; Erent, M.; Thompson, A.I.; Burroughs, N.J.; McAinsh, A.D. Ensemble-level organization of human kinetochores and evidence for distinct tension and attachment sensors. Cell Rep. 2020, 31, 107535. [Google Scholar] [CrossRef]
- Renda, F.; Magidson, V.; Tikhonenko, I.; Fisher, R.; Miles, C.; Mogilner, A.; Khodjakov, A. Effects of malleable kinetochore morphology on measurements of intrakinetochore tension. Open Biol. 2020, 10, 200101. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.S.M.; Botstein, D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 1993, 135, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Biggins, S.; Severin, F.F.; Bhalla, N.; Sassoon, I.; Hyman, A.A.; Murray, A.W. The conserved protein kinase ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999, 13, 532–544. [Google Scholar] [CrossRef]
- Tanaka, T.U.; Rachidi, N.; Janke, C.; Pereira, G.; Galova, M.; Schiebel, E.; Stark, M.J.; Nasmyth, K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 2002, 108, 317–329. [Google Scholar] [CrossRef]
- Kallio, M.J.; McCleland, M.L.; Stukenberg, P.T.; Gorbsky, G.J. Inhibition of aurora b kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 2002, 12, 900–905. [Google Scholar] [CrossRef] [Green Version]
- Cheeseman, I.M.; Anderson, S.; Jwa, M.; Green, E.M.; Kang, J.; Yates, J.R., III; Chan, C.S.; Drubin, D.G.; Barnes, G. Phospho-regulation of kinetochore-microtubule attachments by the aurora kinase Ipl1p. Cell 2002, 111, 163–172. [Google Scholar] [CrossRef] [Green Version]
- Francisco, L.; Wang, W.; Chan, C.S. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol. 1994, 14, 4731. [Google Scholar] [CrossRef] [Green Version]
- Ruchaud, S.; Carmena, M.; Earnshaw, W.C. Chromosomal passengers: Conducting cell division. Nat. Rev. Mol. Cell Biol. 2007, 8, 798–812. [Google Scholar] [CrossRef]
- Abad, M.A.; Ruppert, J.G.; Buzuk, L.; Wear, M.; Zou, J.; Webb, K.M.; Kelly, D.A.; Voigt, P.; Rappsilber, J.; Earnshaw, W.C. Borealin–nucleosome interaction secures chromosome association of the chromosomal passenger complex. J. Cell Biol. 2019, 218, 3912–3925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, A.E.; Ghenoiu, C.; Xue, J.Z.; Zierhut, C.; Kimura, H.; Funabiki, H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic Kinase Aurora B. Science 2010, 330, 235–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.; Zhang, Z.; Chen, Q.; Yan, H.; Zhang, M.; Zhou, L.; Xu, J.; Lu, W.; Wang, F. Centromere-localized Aurora B Kinase is required for the fidelity of chromosome segregation. J. Cell Biol. 2020, 219, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Dai, J.; Daum, J.R.; Niedzialkowska, E.; Banerjee, B.; Stukenberg, P.T.; Gorbsky, G.J.; Higgins, J.M. Histone H3 Thr-3 phosphorylation by haspin positions Aurora B at centromeres in mitosis. Science 2010, 330, 231–235. [Google Scholar] [CrossRef] [Green Version]
- Yue, Z.; Carvalho, A.; Xu, Z.; Yuan, X.; Cardinale, S.; Ribeiro, S.; Lai, F.; Ogawa, H.; Gudmundsdottir, E.; Gassmann, R. Deconstructing survivin: Comprehensive genetic analysis of survivin function by conditional knockout in a vertebrate cell line. J. Cell Biol. 2008, 183, 279–296. [Google Scholar] [CrossRef] [Green Version]
- Vader, G.; Medema, R.H.; Lens, S.M.A. The chromosomal passenger complex: Guiding Aurora-B through mitosis. J. Cell Biol. 2006, 173, 833–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeyaprakash, A.; Basquin, C.; Jayachandran, U.; Conti, E. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure 2011, 19, 1625–1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, R.R.; Wheatley, S.P.; Gouldsworthy, A.M.; Kandels-Lewis, S.E.; Carmena, M.; Smythe, C.; Gerloff, D.L.; Earnshaw, W.C. INCENP binds the Aurora-related Kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000, 10, 1075–1078. [Google Scholar] [CrossRef] [Green Version]
- Bishop, J.D.; Han, Z.; Schumacher, J.M. the caenorhabditis elegans Aurora B Kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol. Biol. Cell 2005, 16, 742–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheatley, S.P.; Kandels-Lewis, S.E.; Adams, R.R.; Ainsztein, A.M.; Earnshaw, W.C. INCENP binds directly to tubulin and requires dynamic microtubules to target to the cleavage furrow. Exp. Cell Res. 2001, 262, 122–127. [Google Scholar] [CrossRef]
- Samejima, K.; Platani, M.; Wolny, M.; Ogawa, H.; Vargiu, G.; Knight, P.J.; Peckham, M.; Earnshaw, W.C. The inner centromere protein (INCENP) coil is a single α-helix (SAH) domain that binds directly to microtubules and is important for chromosome passenger complex (CPC) localization and function in mitosis. J. Biol. Chem. 2015, 290, 21460–21472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandall, S.; Severin, F.; McLeod, I.X.; Yates, J.R., III; Oegema, K.; Hyman, A.; Desai, A. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell 2006, 127, 1179–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haase, J.; Bonner, M.K.; Halas, H.; Kelly, A.E. Distinct roles of the chromosomal passenger complex in the detection of and response to errors in kinetochore-microtubule attachment. Dev. Cell 2017, 42, 640–654. [Google Scholar] [CrossRef] [Green Version]
- Resnick, T.D.; Satinover, D.L.; MacIsaac, F.; Stukenberg, P.T.; Earnshaw, W.C.; Orr-Weaver, T.L.; Carmena, M. INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the shugoshin MEI-S332 in drosophila. Dev. Cell 2006, 11, 57–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuller, B.G.; Lampson, M.A.; Foley, E.A.; Rosasco-Nitcher, S.; Le, K.V.; Tobelmann, P.; Brautigan, D.L.; Stukenberg, P.T.; Kapoor, T.M. Midzone activation of Aurora B in anaphase produces an intracellular phosphorylation gradient. Nature 2008, 453, 1132–1136. [Google Scholar] [CrossRef] [Green Version]
- Afonso, O.; Castellani, C.M.; Cheeseman, L.P.; Ferreira, J.G.; Orr, B.; Ferreira, L.T.; Chambers, J.J.; Morais-de-Sá, E.; Maresca, T.J.; Maiato, H. Spatiotemporal control of mitotic exit during anaphase by an Aurora B-Cdk1 crosstalk. Elife 2019, 8, e47646. [Google Scholar] [CrossRef]
- Hengeveld, R.C.C.; Vromans, M.J.M.; Vleugel, M.; Hadders, M.A.; Lens, S.M.A. Inner Centromere localization of the CPC maintains centromere cohesion and allows mitotic checkpoint silencing. Nat. Commun. 2017, 8, 15542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadders, M.A.; Hindriksen, S.; Truong, M.A.; Mhaskar, A.N.; Wopken, J.P.; Vromans, M.J.; Lens, S. Untangling the contribution of Haspin and Bub1 to Aurora B function during mitosis. J. Cell Biol. 2020, 219, e201907087. [Google Scholar] [CrossRef] [PubMed]
- Broad, A.J.; DeLuca, K.F.; DeLuca, J.G. Aurora B Kinase is recruited to multiple discrete kinetochore and centromere regions in human cells. J. Cell Biol. 2020, 219, e201905144. [Google Scholar] [CrossRef] [PubMed]
- Ciferri, C.; Pasqualato, S.; Screpanti, E.; Varetti, G.; Santaguida, S.; Dos Reis, G.; Maiolica, A.; Polka, J.; De Luca, J.G.; De Wulf, P. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 2008, 133, 427–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, R.R.; Al-Bassam, J.; Harrison, S.C. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 2007, 14, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Cheerambathur, D.K.; Prevo, B.; Hattersley, N.; Lewellyn, L.; Corbett, K.D.; Oegema, K.; Desai, A. Dephosphorylation of the Ndc80 tail stabilizes kinetochore-microtubule attachments via the ska complex. Dev. Cell 2017, 41, 424–437. [Google Scholar] [CrossRef] [Green Version]
- DeLuca, J.G.; Gall, W.E.; Ciferri, C.; Cimini, D.; Musacchio, A.; Salmon, E.D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 2006, 127, 969–982. [Google Scholar] [CrossRef] [Green Version]
- Cheeseman, I.M.; Chappie, J.S.; Wilson-Kubalek, E.M.; Desai, A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 2006, 127, 983–997. [Google Scholar] [CrossRef] [Green Version]
- Bonner, M.K.; Haase, J.; Swinderman, J.; Halas, H.; Miller Jenkins, L.M.; Kelly, A.E. Enrichment of Aurora B Kinase at the inner kinetochore controls outer kinetochore assembly. J. Cell Biol. 2019, 218, 3237–3257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Zheng, F.; Wang, C.; Wu, M.; Zhang, X.; Wang, Q.; Yao, X.; Fu, C.; Zhang, X.; Zang, J. Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis. Proc. Natl. Acad. Sci. USA 2017, 114, E10667–E10676. [Google Scholar] [CrossRef] [Green Version]
- Lan, W.; Zhang, X.; Kline-Smith, S.L.; Rosasco, S.E.; Barrett-Wilt, G.A.; Shabanowitz, J.; Hunt, D.F.; Walczak, C.E.; Stukenberg, P.T. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 2004, 14, 273–286. [Google Scholar] [CrossRef]
- Andrews, P.D.; Ovechkina, Y.; Morrice, N.; Wagenbach, M.; Duncan, K.; Wordeman, L.; Swedlow, J.R. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell 2004, 6, 253–268. [Google Scholar] [CrossRef] [Green Version]
- McHugh, T.; Zou, J.; Volkov, V.A.; Bertin, A.; Talapatra, S.K.; Rappsilber, J.; Dogterom, M.; Welburn, J.P. The depolymerase activity of MCAK shows a graded response to Aurora B Kinase phosphorylation through allosteric regulation. J. Cell. Sci. 2019, 132, jcs228353. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, Y.; Cormier, A.; Tyers, R.G.; Pigula, A.; Peng, Y.; Drubin, D.G.; Barnes, G. Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP regulates CPC–spindle Interaction to ensure proper microtubule dynamics. J. Cell Biol. 2011, 194, 137–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, J.D.; Schumacher, J.M. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase Stimulates Aurora B kinase activity. J. Biol. Chem. 2002, 277, 27577–27580. [Google Scholar] [CrossRef] [Green Version]
- Honda, R.; Korner, R.; Nigg, E.A. Exploring the Functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell 2003, 14, 3325–3341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salimian, K.; Ballister, E.; Smoak, E.; Wood, S.; Panchenko, T.; Lampson, M.; Black, B. Feedback control in sensing chromosome biorientation by the Aurora B Kinase. Curr. Biol. 2011, 21, 1158–1165. [Google Scholar] [CrossRef] [Green Version]
- Yasui, Y.; Urano, T.; Kawajiri, A.; Nagata, K.; Tatsuka, M.; Saya, H.; Furukawa, K.; Takahashi, T.; Izawa, I.; Inagaki, M. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J. Biol. Chem. 2004, 279, 12997–13003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, M.; Goshima, T.; Matsuo, H.; Johmura, Y.; Haruta, M.; Murata, K.; Tanaka, H.; Ikawa, M.; Nakanishi, K.; Nakanishi, M. Essential role of autoactivation circuitry on Aurora B-mediated H2AX-pS121 in mitosis. Nat. Commun. 2016, 7, 12059. [Google Scholar] [CrossRef] [Green Version]
- Ditchfield, C.; Johnson, V.L.; Tighe, A.; Ellston, R.; Haworth, C.; Johnson, T.; Mortlock, A.; Keen, N.; Taylor, S.S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003, 161, 267–280. [Google Scholar] [CrossRef]
- Vigneron, S.; Prieto, S.; Bernis, C.; Labbé, J.; Castro, A.; Lorca, T. Kinetochore localization of spindle checkpoint proteins: Who controls whom? Mol. Biol. Cell 2004, 15, 4584–4596. [Google Scholar] [CrossRef] [Green Version]
- Gurden, M.D.; Anderhub, S.J.; Faisal, A.; Linardopoulos, S. Aurora B prevents premature removal of spindle assembly checkpoint proteins from the kinetochore: A key role for Aurora B in mitosis. Oncotarget 2018, 9, 19525. [Google Scholar] [CrossRef] [Green Version]
- Krenn, V.; Musacchio, A. The Aurora B Kinase in chromosome bi-orientation and spindle checkpoint signaling. Front. Oncol. 2015, 5, 225. [Google Scholar] [CrossRef] [Green Version]
- García-Rodríguez, L.J.; Kasciukovic, T.; Denninger, V.; Tanaka, T.U. Aurora B-INCENP localization at centromeres/inner kinetochores is required for chromosome bi-orientation in budding yeast. Curr. Biol. 2019, 29, 1536–1544. [Google Scholar] [CrossRef] [Green Version]
- Campbell, C.S.; Desai, A. Tension sensing by Aurora B Kinase is independent of survivin-based centromere localization. Nature 2013, 497, 118–121. [Google Scholar] [CrossRef] [Green Version]
- Wheelock, M.S.; Wynne, D.J.; Tseng, B.S.; Funabiki, H. Dual recognition of chromatin and microtubules by INCENP is important for mitotic progression. J. Cell Biol. 2017, 216, 925–941. [Google Scholar] [CrossRef] [Green Version]
- Lampson, M.A.; Cheeseman, I.M. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011, 21, 133–140. [Google Scholar] [CrossRef] [Green Version]
- Beardmore, V.A.; Ahonen, L.J.; Gorbsky, G.J.; Kallio, M.J. Survivin dynamics increases at centromeres during G2/M phase transition and is regulated by microtubule-attachment and Aurora B Kinase activity. J. Cell Sci. 2004, 117, 4033–4042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahonen, L.J.; Kukkonen, A.M.; Pouwels, J.; Bolton, M.A.; Jingle, C.D.; Stukenberg, P.T.; Kallio, M.J. Perturbation of incenp function impedes anaphase chromatid movements and chromosomal passenger protein flux at centromeres. Chromosoma 2009, 118, 71–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.; Ballister, E.R.; Lampson, M.A. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J. Cell Biol. 2011, 194, 539–549. [Google Scholar] [CrossRef]
- Tan, L.; Kapoor, T.M. Examining the dynamics of chromosomal passenger complex (CPC)-dependent phosphorylation during cell division. Proc. Natl. Acad. Sci. USA 2011, 108, 16675–16680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumann, C.; Körner, R.; Hofmann, K.; Nigg, E.A. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 2007, 128, 101–114. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.U. Bi-orienting chromosomes: Acrobatics on the mitotic spindle. Chromosoma 2008, 117, 521–533. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, C.F.; Huttner, D.; Bizard, A.H.; Hirano, S.; Li, T.; Palmai-Pallag, T.; Bjerregaard, V.A.; Liu, Y.; Nigg, E.A.; Wang, L.H. PICH promotes sister chromatid disjunction and co-operates with topoisomerase ii in mitosis. Nat. Commun. 2015, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Craske, B.; Welburn, J.P. Leaving no-one behind: How CENP-E facilitates chromosome alignment. Essays Biochem. 2020, 64, 313–324. [Google Scholar] [PubMed]
- Trinkle-Mulcahy, L.; Andrews, P.D.; Wickramasinghe, S.; Sleeman, J.; Prescott, A.; Lam, Y.W.; Lyon, C.; Swedlow, J.R.; Lamond, A.I. Time-lapse imaging reveals dynamic relocalization of PP1γ throughout the mammalian cell cycle. Mol. Biol. Cell 2003, 14, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Foley, E.A.; Maldonado, M.; Kapoor, T.M. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 2011, 13, 1265–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertz, E.P.T.; Kruse, T.; Davey, N.E.; López-Méndez, B.; Sigurðsson, J.O.; Montoya, G.; Olsen, J.V.; Nilsson, J. A conserved motif provides binding specificity to the PP2A-B56 phosphatase. Mol. Cell 2016, 63, 686–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eshleman, H.D.; Morgan, D.O. Sgo1 recruits PP2A to chromosomes to ensure sister chromatid bi-orientation during mitosis. J. Cell Sci. 2014, 127, 4974–4983. [Google Scholar] [CrossRef] [Green Version]
- Meppelink, A.; Kabeche, L.; Vromans, M.J.; Compton, D.A.; Lens, S.M. Shugoshin-1 balances Aurora B Kinase activity via PP2A to promote chromosome bi-orientation. Cell Rep. 2015, 11, 508–515. [Google Scholar] [CrossRef] [Green Version]
- Dohadwala, M.; e Silva, E.D.C.; Hall, F.L.; Williams, R.T.; Carbonaro-Hall, D.A.; Nairn, A.C.; Greengard, P.; Berndt, N. Phosphorylation and inactivation of protein phosphatase 1 by cyclin-dependent kinases. Proc. Natl. Acad. Sci. USA 1994, 91, 6408–6412. [Google Scholar] [CrossRef] [Green Version]
- Yamano, H.; Ishii, K.; Yanagida, M. Phosphorylation of dis2 protein phosphatase at the C-terminal cdc2 consensus and its potential role in cell cycle regulation. EMBO J. 1994, 13, 5310–5318. [Google Scholar] [CrossRef]
- Grallert, A.; Boke, E.; Hagting, A.; Hodgson, B.; Connolly, Y.; Griffiths, J.R.; Smith, D.L.; Pines, J.; Hagan, I.M. A PP1–PP2A phosphatase relay controls mitotic progression. Nature 2015, 517, 94–98. [Google Scholar] [CrossRef] [Green Version]
- Asai, Y.; Fukuchi, K.; Tanno, Y.; Koitabashi-Kiyozuka, S.; Kiyozuka, T.; Noda, Y.; Matsumura, R.; Koizumi, T.; Watanabe, A.; Nagata, K. Aurora B Kinase activity is regulated by SET/TAF1 on Sgo2 at the inner centromere. J. Cell Biol. 2019, 218, 3223–3236. [Google Scholar] [CrossRef] [Green Version]
- Asai, Y.; Matsumura, R.; Hasumi, Y.; Susumu, H.; Nagata, K.; Watanabe, Y.; Terada, Y. SET/TAF1 forms a distance-dependent feedback loop with Aurora B and Bub1 as a tension sensor at centromeres. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Roy, B.; Verma, V.; Sim, J.; Fontan, A.; Joglekar, A.P. Delineating the contribution of Spc105-bound PP1 to spindle checkpoint silencing and kinetochore microtubule attachment regulation. J. Cell Biol. 2019, 218, 3926–3942. [Google Scholar] [CrossRef]
- Petsalaki, E.; Akoumianaki, T.; Black, E.J.; Gillespie, D.A.; Zachos, G. Phosphorylation at Serine 331 is Required for Aurora B Activation. J. Cell Biol. 2011, 195, 449–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petsalaki, E.; Zachos, G. Chk2 Prevents Mitotic Exit when the Majority of Kinetochores are Unattached. J. Cell Biol. 2014, 205, 339–356. [Google Scholar] [CrossRef] [PubMed]
- Caldas, G.V.; DeLuca, K.F.; DeLuca, J.G. KNL1 facilitates phosphorylation of outer kinetochore proteins by promoting Aurora B Kinase activity. J. Cell Biol. 2013, 203, 957–969. [Google Scholar] [CrossRef]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef] [Green Version]
- Sarangapani, K.K.; Asbury, C.L. Catch and release: How do kinetochores hook the right microtubules during mitosis? Trends Genet. 2014, 30, 150–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, A.F.; Udy, D.B.; Dumont, S. Hec1 tail phosphorylation differentially regulates mammalian kinetochore coupling to polymerizing and depolymerizing microtubules. Curr. Biol. 2017, 27, 1692–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armond, J.W.; Harry, E.F.; McAinsh, A.D.; Burroughs, N.J. Inferring the forces controlling metaphase kinetochore oscillations by reverse engineering system dynamics. PLoS Comput. Biol. 2015, 11, e1004607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banigan, E.J.; Chiou, K.K.; Ballister, E.R.; Mayo, A.M.; Lampson, M.A.; Liu, A.J. Minimal model for collective kinetochore–microtubule dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 12699–12704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, A. Shake it Off: The elimination of erroneous kinetochore-microtubule attachments and chromosome oscillation. Int. J. Mol. Sci. 2021, 22, 3174. [Google Scholar] [CrossRef]
- Iemura, K.; Natsume, T.; Maehara, K.; Kanemaki, M.T.; Tanaka, K. Chromosome oscillation promotes Aurora A–dependent Hec1 phosphorylation and mitotic fidelity. J. Cell Biol. 2021, 220, e202006116. [Google Scholar] [CrossRef]
- Wakiya, M.; Nishi, E.; Kawai, S.; Yamada, K.; Katsumata, K.; Hirayasu, A.; Itabashi, Y.; Yamamoto, A. Chiasmata and the kinetochore component Dam1 are crucial for elimination of erroneous chromosome attachments and centromere oscillation at meiosis I. Open Biol. 2021, 11, 200308. [Google Scholar] [CrossRef]
- Magnaghi-Jaulin, L.; Eot-Houllier, G.; Gallaud, E.; Giet, R. Aurora A protein kinase: To the centrosome and beyond. Biomolecules 2019, 9, 28. [Google Scholar] [CrossRef] [Green Version]
- Eot-Houllier, G.; Magnaghi-Jaulin, L.; Fulcrand, G.; Moyroud, F.; Monier, S.; Jaulin, C. Aurora A-dependent CENP-A phosphorylation at inner centromeres protects bioriented chromosomes against cohesion fatigue. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McVey, S.L.; Cosby, J.K.; Nannas, N.J. Aurora B Tension Sensing Mechanisms in the Kinetochore Ensure Accurate Chromosome Segregation. Int. J. Mol. Sci. 2021, 22, 8818. https://doi.org/10.3390/ijms22168818
McVey SL, Cosby JK, Nannas NJ. Aurora B Tension Sensing Mechanisms in the Kinetochore Ensure Accurate Chromosome Segregation. International Journal of Molecular Sciences. 2021; 22(16):8818. https://doi.org/10.3390/ijms22168818
Chicago/Turabian StyleMcVey, Shelby L., Jenna K. Cosby, and Natalie J. Nannas. 2021. "Aurora B Tension Sensing Mechanisms in the Kinetochore Ensure Accurate Chromosome Segregation" International Journal of Molecular Sciences 22, no. 16: 8818. https://doi.org/10.3390/ijms22168818